Archaeal RNA polymerase: the influence of the protruding stalk in crystal packing and preliminary biophysical analysis of the Rpo13 subunit.
Wojtas, M., Peralta, B., Ondiviela, M., Mogni, M., Bell, S.D., Abrescia, N.G.(2011) Biochem Soc Trans 39: 25-30
- PubMed: 21265742
- DOI: https://doi.org/10.1042/BST0390025
- Primary Citation of Related Structures:
2Y0S - PubMed Abstract:
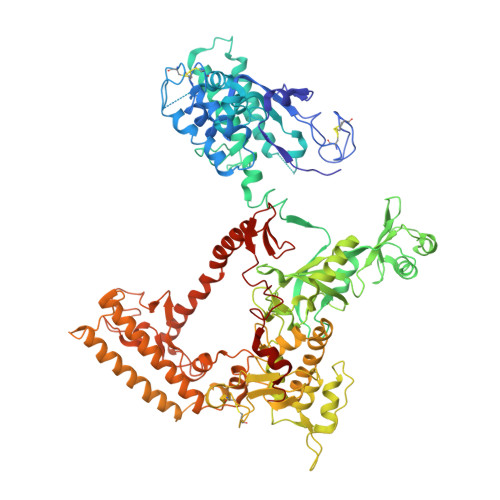
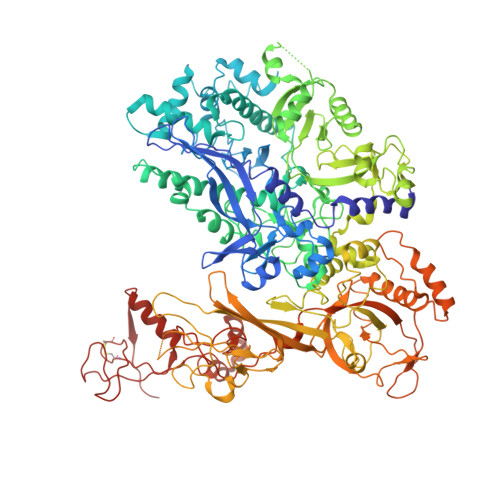
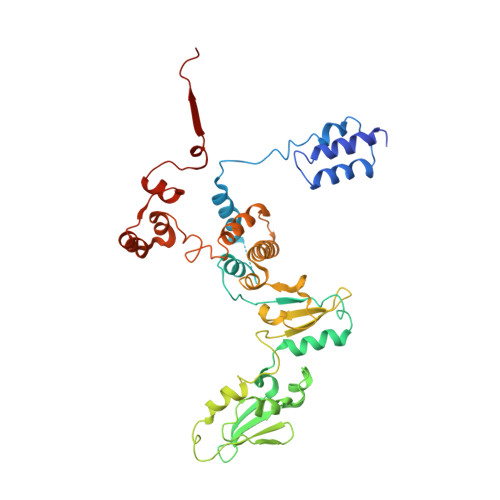
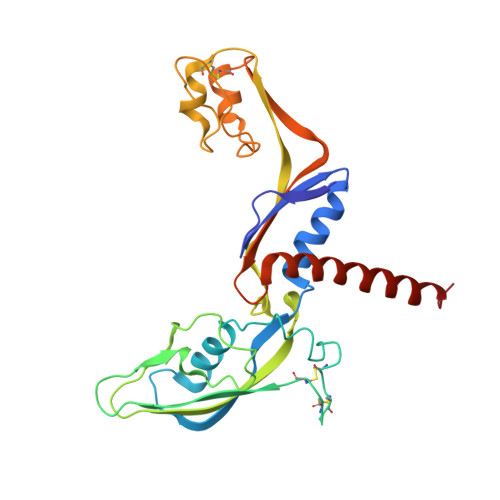
We review recent results on the complete structure of the archaeal RNAP (RNA polymerase) enzyme of Sulfolobus shibatae. We compare the three crystal forms in which this RNAP packs (space groups P2₁2₁2₁, P2₁2₁2 and P2₁) and provide a preliminary biophysical characterization of the newly identified 13-subunit Rpo13. The availability of different crystal forms for this RNAP allows the analysis of the packing degeneracy and the intermolecular interactions that determine this degeneracy. We observe the pivotal role played by the protruding stalk composed of subunits Rpo4 and Rpo7 in the lattice contacts. Aided by MALLS (multi-angle laser light scattering), we have initiated the biophysical characterization of the recombinantly expressed and purified subunit Rpo13, a necessary step towards the understanding of Rpo13's role in archaeal transcription.
Organizational Affiliation:
Structural Biology Unit, CIC bioGUNE, CIBERehd, 48160 Derio, Spain.