A P-loop Mutation in Galpha Subunits Prevents Transition to the Active State: Implications for G-protein Signaling in Fungal Pathogenesis
Bosch, D.E., Willard, F.S., Ramanujam, R., Kimple, A.J., Willard, M.D., Naqvi, N.I., Siderovski, D.P.(2012) PLoS Pathog 8: e1002553-e1002553
- PubMed: 22383884
- DOI: https://doi.org/10.1371/journal.ppat.1002553
- Primary Citation of Related Structures:
3QE0, 3QI2 - PubMed Abstract:
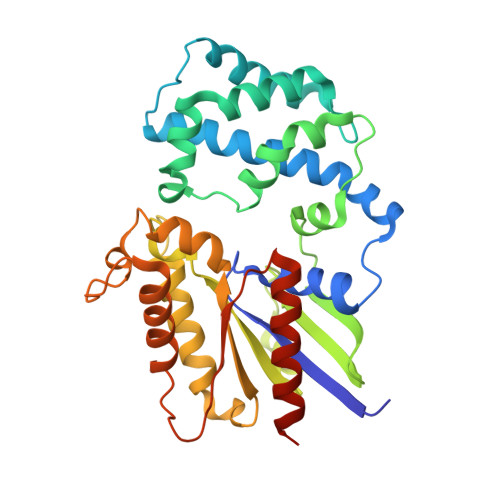
Heterotrimeric G-proteins are molecular switches integral to a panoply of different physiological responses that many organisms make to environmental cues. The switch from inactive to active Gαβγ heterotrimer relies on nucleotide cycling by the Gα subunit: exchange of GTP for GDP activates Gα, whereas its intrinsic enzymatic activity catalyzes GTP hydrolysis to GDP and inorganic phosphate, thereby reverting Gα to its inactive state. In several genetic studies of filamentous fungi, such as the rice blast fungus Magnaporthe oryzae, a G42R mutation in the phosphate-binding loop of Gα subunits is assumed to be GTPase-deficient and thus constitutively active. Here, we demonstrate that Gα(G42R) mutants are not GTPase deficient, but rather incapable of achieving the activated conformation. Two crystal structure models suggest that Arg-42 prevents a typical switch region conformational change upon Gα(i1)(G42R) binding to GDP·AlF(4)(-) or GTP, but rotameric flexibility at this locus allows for unperturbed GTP hydrolysis. Gα(G42R) mutants do not engage the active state-selective peptide KB-1753 nor RGS domains with high affinity, but instead favor interaction with Gβγ and GoLoco motifs in any nucleotide state. The corresponding Gα(q)(G48R) mutant is not constitutively active in cells and responds poorly to aluminum tetrafluoride activation. Comparative analyses of M. oryzae strains harboring either G42R or GTPase-deficient Q/L mutations in the Gα subunits MagA or MagB illustrate functional differences in environmental cue processing and intracellular signaling outcomes between these two Gα mutants, thus demonstrating the in vivo functional divergence of G42R and activating G-protein mutants.
Organizational Affiliation:
Department of Pharmacology, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.