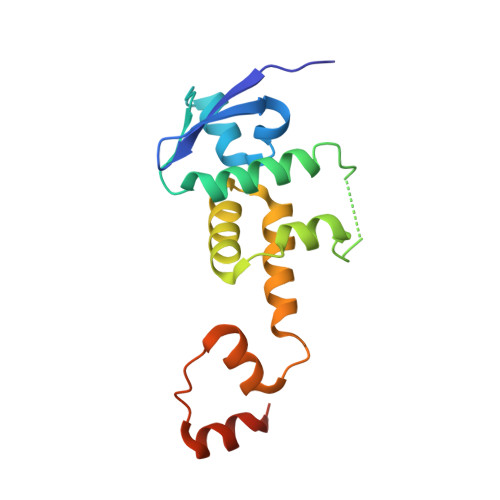
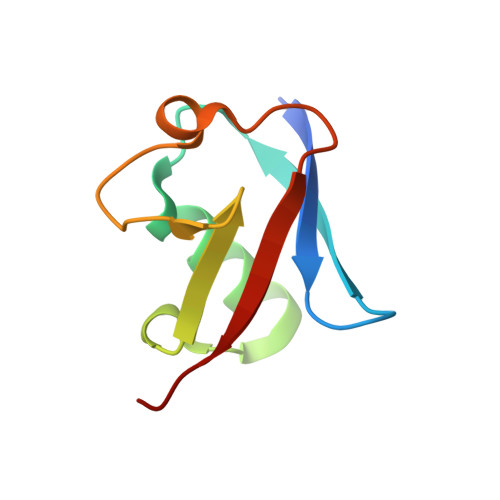
Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1-F-box interface.
Gorelik, M., Orlicky, S., Sartori, M.A., Tang, X., Marcon, E., Kurinov, I., Greenblatt, J.F., Tyers, M., Moffat, J., Sicheri, F., Sidhu, S.S.(2016) Proc Natl Acad Sci U S A 113: 3527-3532
- PubMed: 26976582
- DOI: https://doi.org/10.1073/pnas.1519389113
- Primary Citation of Related Structures:
5IBK - PubMed Abstract:
Skp1-Cul1-F-box (SCF) E3 ligases play key roles in multiple cellular processes through ubiquitination and subsequent degradation of substrate proteins. Although Skp1 and Cul1 are invariant components of all SCF complexes, the 69 different human F-box proteins are variable substrate binding modules that determine specificity. SCF E3 ligases are activated in many cancers and inhibitors could have therapeutic potential. Here, we used phage display to develop specific ubiquitin-based inhibitors against two F-box proteins, Fbw7 and Fbw11. Unexpectedly, the ubiquitin variants bind at the interface of Skp1 and F-box proteins and inhibit ligase activity by preventing Cul1 binding to the same surface. Using structure-based design and phage display, we modified the initial inhibitors to generate broad-spectrum inhibitors that targeted many SCF ligases, or conversely, a highly specific inhibitor that discriminated between even the close homologs Fbw11 and Fbw1. We propose that most F-box proteins can be targeted by this approach for basic research and for potential cancer therapies.
Organizational Affiliation:
Banting and Best Department of Medical Research, Terrence Donnelly Center for Cellular and Biomolecular Research, University of Toronto, Toronto, ON, Canada M5S 3E1; Department of Molecular Genetics, Terrence Donnelly Center for Cellular and Biomolecular Research, University of Toronto, Toronto, ON, Canada M5S 3E1;