The murine orthologue of human antichymotrypsin: a structural paradigm for clade A3 serpins.
Horvath, A.J., Irving, J.A., Rossjohn, J., Law, R.H., Bottomley, S.P., Quinsey, N.S., Pike, R.N., Coughlin, P.B., Whisstock, J.C.(2005) J Biol Chem 280: 43168-43178
- PubMed: 16141197
- DOI: https://doi.org/10.1074/jbc.M505598200
- Primary Citation of Related Structures:
1YXA - PubMed Abstract:
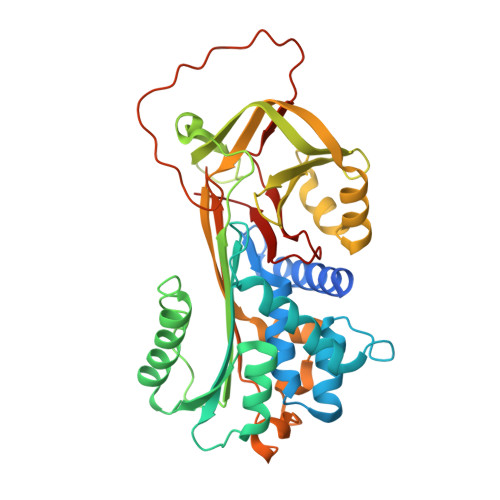
Antichymotrypsin (SERPINA3) is a widely expressed member of the serpin superfamily, required for the regulation of leukocyte proteases released during an inflammatory response and with a permissive role in the development of amyloid encephalopathy. Despite its biological significance, there is at present no available structure of this serpin in its native, inhibitory state. We present here the first fully refined structure of a murine antichymotrypsin orthologue to 2.1 A, which we propose as a template for other antichymotrypsin-like serpins. A most unexpected feature of the structure of murine serpina3n is that it reveals the reactive center loop (RCL) to be partially inserted into the A beta-sheet, a structural motif associated with ligand-dependent activation in other serpins. The RCL is, in addition, stabilized by salt bridges, and its plane is oriented at 90 degrees to the RCL of antitrypsin. A biochemical and biophysical analysis of this serpin demonstrates that it is a fast and efficient inhibitor of human leukocyte elastase (ka: 4 +/- 0.9 x 10(6) m(-1) s(-)1) and cathepsin G (ka: 7.9 +/- 0.9 x 10(5) m(-1) s(-)1) giving a spectrum of activity intermediate between that of human antichymotrypsin and human antitrypsin. An evolutionary analysis reveals that residues subject to positive selection and that have contributed to the diversity of sequences in this sub-branch (A3) of the serpin superfamily are essentially restricted to the P4-P6' region of the RCL, the distal hinge, and the loop between strands 4B and 5B.
Organizational Affiliation:
Australian Centre for Blood Diseases, Monash University, Commercial Road, 6th Floor, MacFarlane Burnet Building, Alfred Medical Research Precinct, Prahran, Victoria, 3181.