Recombinant production and solution structure of lipid transfer protein from lentil Lens culinaris.
Gizatullina, A.K., Finkina, E.I., Mineev, K.S., Melnikova, D.N., Bogdanov, I.V., Telezhinskaya, I.N., Balandin, S.V., Shenkarev, Z.O., Arseniev, A.S., Ovchinnikova, T.V.(2013) Biochem Biophys Res Commun 439: 427-432
- PubMed: 23998937
- DOI: https://doi.org/10.1016/j.bbrc.2013.08.078
- Primary Citation of Related Structures:
2MAL - PubMed Abstract:
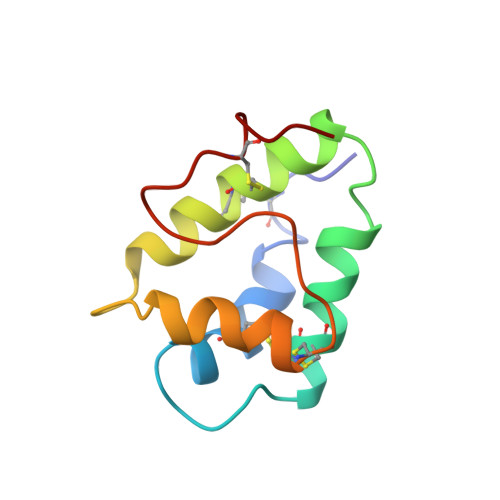
Lipid transfer protein, designated as Lc-LTP2, was isolated from seeds of the lentil Lens culinaris. The protein has molecular mass 9282.7Da, consists of 93 amino acid residues including 8 cysteines forming 4 disulfide bonds. Lc-LTP2 and its stable isotope labeled analogues were overexpressed in Escherichia coli and purified. Antimicrobial activity of the recombinant protein was examined, and its spatial structure was studied by NMR spectroscopy. The polypeptide chain of Lc-LTP2 forms four α-helices (Cys4-Leu18, Pro26-Ala37, Thr42-Ala56, Thr64-Lys73) and a long C-terminal tail without regular secondary structure. Side chains of the hydrophobic residues form a relatively large internal tunnel-like lipid-binding cavity (van der Waals volume comes up to ∼600Å(3)). The side-chains of Arg45, Pro79, and Tyr80 are located near an assumed mouth of the cavity. Titration with dimyristoyl phosphatidylglycerol (DMPG) revealed formation of the Lc-LTP2/lipid non-covalent complex accompanied by rearrangements in the protein spatial structure and expansion of the internal cavity. The resultant Lc-LTP2/DMPG complex demonstrates limited lifetime and dissociates within tens of hours.
Organizational Affiliation:
Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Miklukho-Maklaya str., 16/10, 117997 Moscow, Russia; Moscow Institute of Physics and Technology (State University), Department of Physicochemical Biology and Biotechnology, Institutskii per., 9, 141700, Dolgoprudny, Moscow Region, Russia.