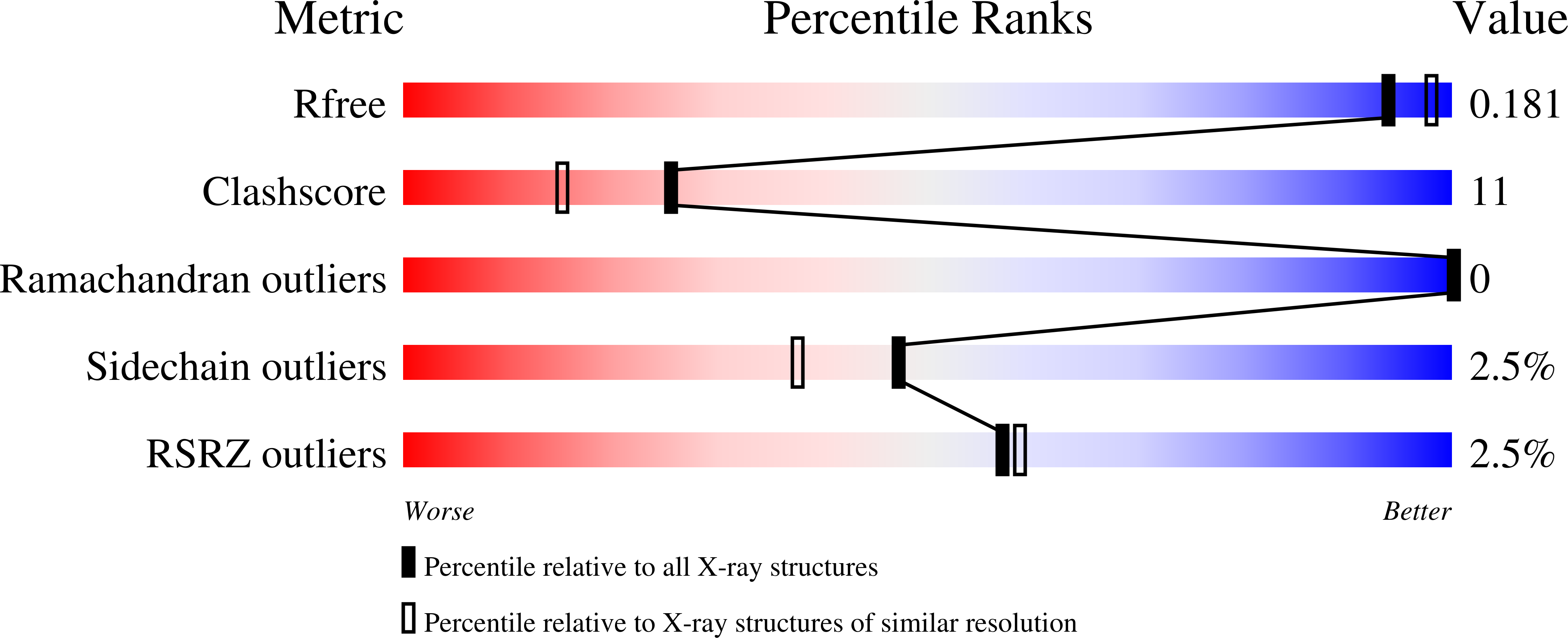
Structure of the His269Arg mutant of the rat aldose reductase-like protein AKR1B14 complexed with NADPH.
Sundaram, K., Endo, S., Matsunaga, T., Tanaka, N., Hara, A., El-Kabbani, O.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 400-403
- PubMed: 22505406
- DOI: https://doi.org/10.1107/S1744309112008810
- Primary Citation of Related Structures:
3QKZ - PubMed Abstract:
Rat aldose reductase-like protein (AKR1B14) is an orthologue of mouse vas deferens protein (AKR1B7) and plays roles in the detoxification of reactive aldehydes and synthesis of prostaglandin F(2α). Here, the 1.87 Å resolution crystal structure of the His269Arg mutant of AKR1B14 complexed with NADPH is described and shows that the negatively charged 2'-phosphate group of the coenzyme forms an ionic interaction with the positively charged guanidinium group of Arg269 that is also observed in the human aldose reductase (AKR1B1) structure. Previous experiments on the site-directed mutagenesis of His269 to Arg, Phe and Met revealed fourfold, sevenfold and 127-fold increases in the K(m) for NADPH, respectively, which are in agreement with the present molecular-modelling and X-ray crystallographic studies. This is the first tertiary structure of a mutant form of this AKR1B7 orthologue to be reported in order to investigate the structure-function relationship of the nonconserved His269 and its role in coenzyme binding.
Organizational Affiliation:
Medicinal Chemistry and Drug Action, Monash Institute of Pharmaceutical Sciences, Monash University, 381 Royal Parade, Parkville, Victoria 3052, Australia.