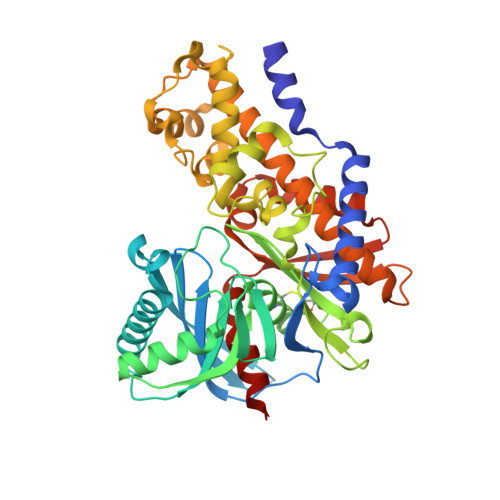
Matched triplicate design sets in the optimisation of glucokinase activators maximising medicinal chemistry information content
Waring, M.J., Bennett, S.N.L., Boyd, S., Campbell, L., Davies, R.D.M., Gerhardt, S., Hargreaves, D., G Martin, N., Robb, G.R., Wilkinson, G.To be published.