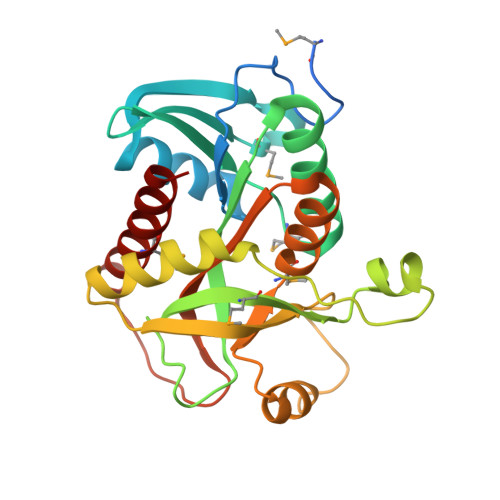
Crystal structure of purine nucleoside phosphorylase from Leptotrichia buccalis C-1013-b, NYSGRC Target 029767.
Malashkevich, V.N., Bhosle, R., Toro, R., Hillerich, B., Gizzi, A., Garforth, S., Kar, A., Chan, M.K., Lafluer, J., Patel, H., Matikainen, B., Chamala, S., Lim, S., Celikgil, A., Villegas, G., Evans, B., Love, J., Fiser, A., Khafizov, K., Seidel, R., Bonanno, J.B., Almo, S.C.To be published.