Structural basis for the 4'-hydroxylation of diclofenac by a microbial cytochrome P450 monooxygenase.
Xu, L.H., Ikeda, H., Liu, L., Arakawa, T., Wakagi, T., Shoun, H., Fushinobu, S.(2015) Appl Microbiol Biotechnol 99: 3081-3091
- PubMed: 25341403
- DOI: https://doi.org/10.1007/s00253-014-6148-y
- Primary Citation of Related Structures:
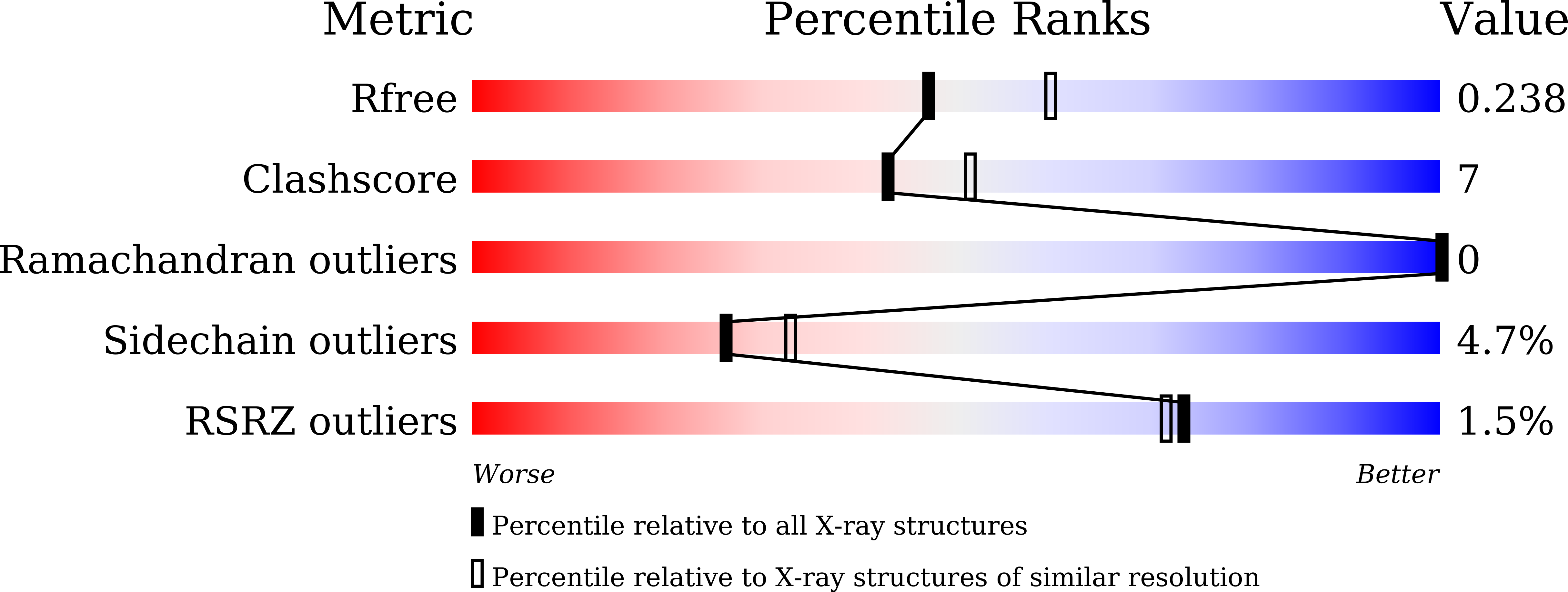
4UBS - PubMed Abstract:
Diclofenac is a nonsteroidal anti-inflammatory drug. It undergoes hydroxylation by mammalian cytochrome P450 enzymes at 4'- and/or 5'-positions. A bacterial P450 enzyme, CYP105D7 from Streptomyces avermitilis, has been shown to catalyze hydroxylation of 1-deoxypentalenic acid and an isoflavone daidzein. Here, we demonstrated that CYP105D7 also catalyzes hydroxylation of diclofenac at the C4'-position. A spectroscopic analysis showed that CYP105D7 binds diclofenac in a slightly cooperative manner with an affinity of 65 μM and a Hill coefficient of 1.16. The crystal structure of CYP105D7 in complex with diclofenac was determined at 2.2 Å resolution. The distal pocket of CYP105D7 contains two diclofenac molecules, illustrating drug recognition with a double-ligand-binding mode. The C3' and C4' atoms of the dichlorophenyl ring of one diclofenac molecule are positioned near the heme iron, suggesting that it is positioned appropriately for aromatic hydroxylation to yield the 4'-hydroxylated product. However, recognition of diclofenac by CYP105D7 was completely different from that of rabbit CYP2C5, which binds one diclofenac molecule with a cluster of water molecules. The distal pocket of CYP105D7 contains four arginine residues, forming a wall of the substrate-binding pocket, and the arginine residues are conserved in bacterial P450s in the CYP105 family.
Organizational Affiliation:
Ocean College, Zhejiang University, 866 Yuhangtang Road, Hangzhou, Zhejiang, 310058, China, lianhuaxu@zju.edu.cn.