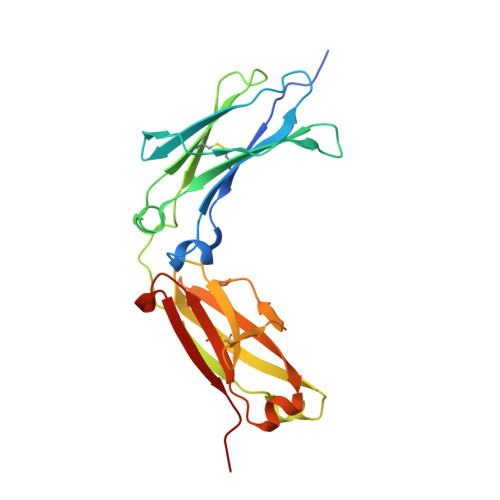
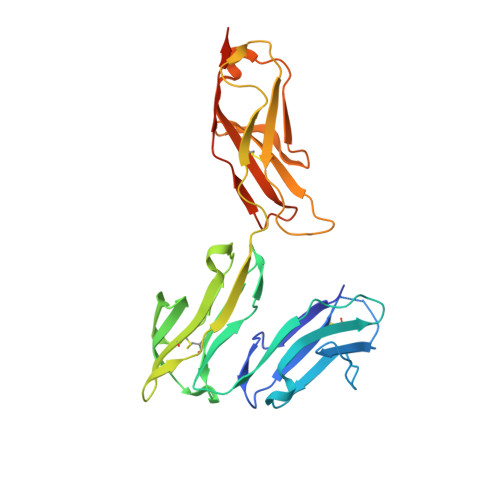
Structural basis for binding of human IgG1 to its high-affinity human receptor Fc gamma RI
Kiyoshi, M., Caaveiro, J.M.M., Kawai, T., Tashiro, S., Ide, T., Asaoka, Y., Hatayama, K., Tsumoto, K.(2015) Nat Commun 6: 6866-6866
- PubMed: 25925696
- DOI: https://doi.org/10.1038/ncomms7866
- Primary Citation of Related Structures:
4W4N, 4W4O - PubMed Abstract:
Cell-surface Fcγ receptors mediate innate and adaptive immune responses. Human Fcγ receptor I (hFcγRI) binds IgGs with high affinity and is the only Fcγ receptor that can effectively capture monomeric IgGs. However, the molecular basis of hFcγRI's interaction with Fc has not been determined, limiting our understanding of this major immune receptor. Here we report the crystal structure of a complex between hFcγRI and human Fc, at 1.80 Å resolution, revealing an unique hydrophobic pocket at the surface of hFcγRI perfectly suited for residue Leu235 of Fc, which explains the high affinity of this complex. Structural, kinetic and thermodynamic data demonstrate that the binding mechanism is governed by a combination of non-covalent interactions, bridging water molecules and the dynamic features of Fc. In addition, the hinge region of hFcγRI-bound Fc adopts a straight conformation, potentially orienting the Fab moiety. These findings will stimulate the development of novel therapeutic strategies involving hFcγRI.
Organizational Affiliation:
Department of Bioengineering, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Tokyo 113-8656, Japan.