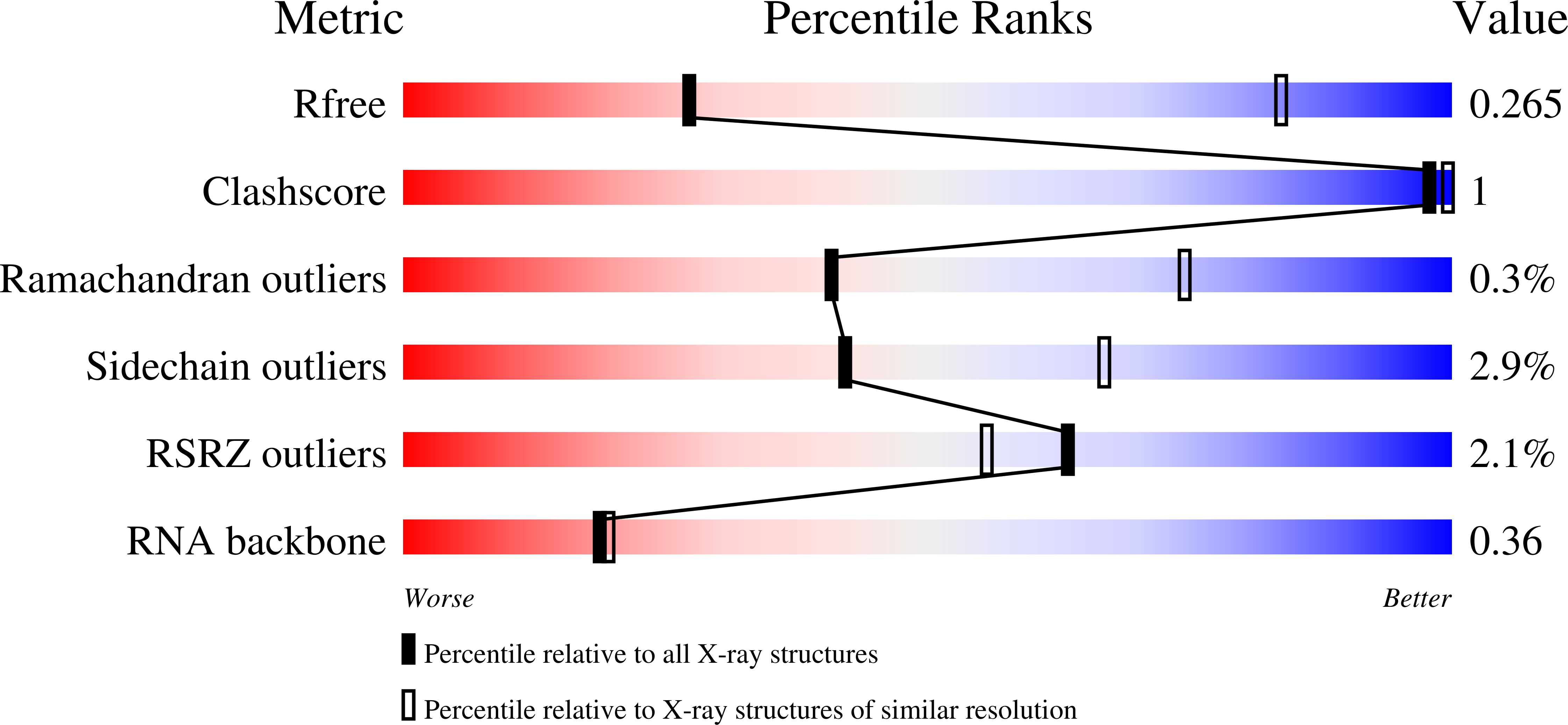
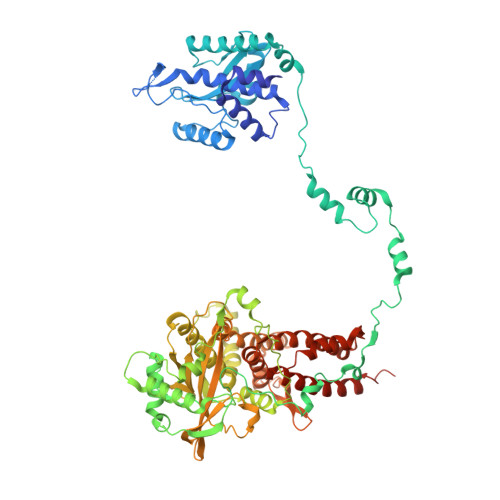
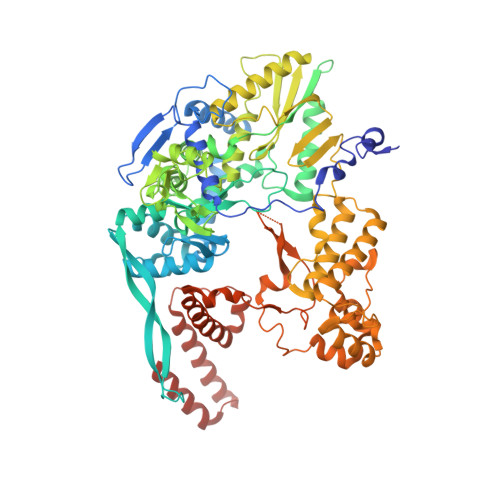
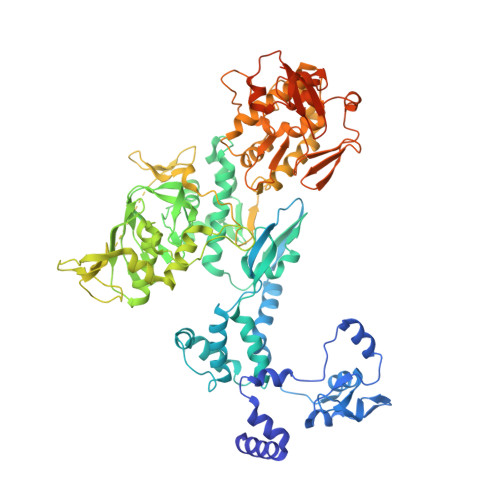
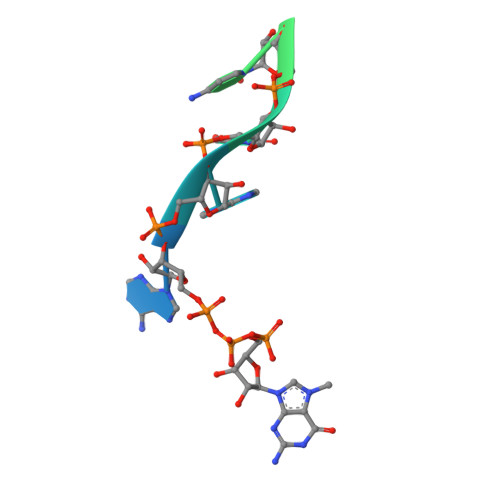
An in vitro fluorescence based study of initiation of RNA synthesis by influenza B polymerase.
Reich, S., Guilligay, D., Cusack, S.(2017) Nucleic Acids Res 45: 3353-3368
- PubMed: 28126917
- DOI: https://doi.org/10.1093/nar/gkx043
- Primary Citation of Related Structures:
5MSG - PubMed Abstract:
Influenza polymerase replicates, via a complementary RNA intermediate (cRNA), and transcribes the eight viral RNA (vRNA) genome segments. To initiate RNA synthesis it is bound to the conserved 5΄ and 3΄ extremities of the vRNA or cRNA (the 'promoter'). 5΄-3΄ base-pairing in the distal promoter region is essential to position the template RNA at the polymerase active site, as shown by a new crystal structure with the 3΄ end threading through the template entry tunnel. We develop fluorescence polarization assays to quantify initiation of cap-primed (transcription) or unprimed (replication) RNA synthesis by recombinant influenza B polymerase bound to the vRNA or cRNA promoter. The rate-limiting step is formation of a primed initiation complex with minimally ApG required to stabilize the 3΄ end of the template within the active-site. Polymerase bound to the vRNA promoter initiates RNA synthesis terminally, while the cRNA promoter directs internal initiation at a significantly lower rate. Progression to elongation requires breaking the promoter 5΄-3΄ base-pairing region and favourable compensation by the emerging template-product base-pairs. The RNA synthesis assay is adaptable to high-throughput screening for polymerase inhibitors. In a pilot study, we find that initiation at the cRNA promoter is unusually susceptible to inhibition by 2΄F-2΄dNTPs.
Organizational Affiliation:
Grenoble Outstation, European Molecular Biology Laboratory, Grenoble 38042, France.