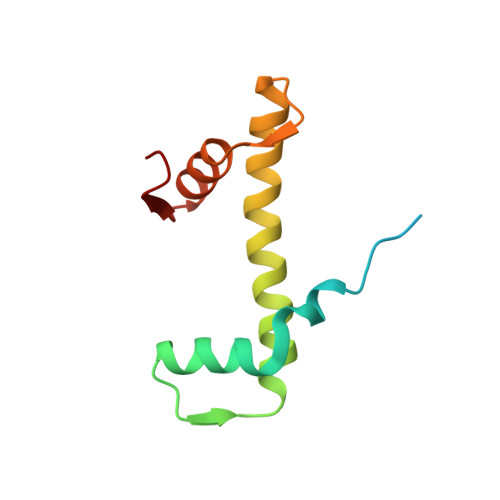
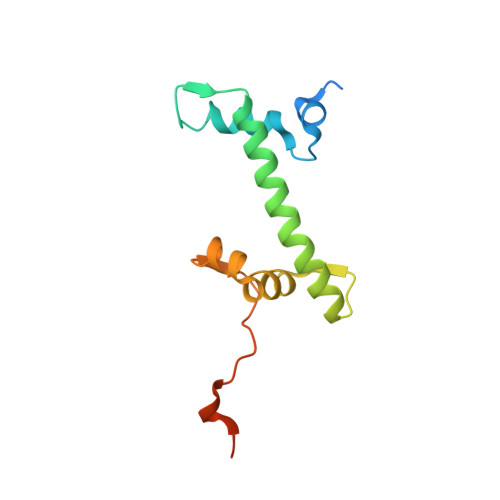
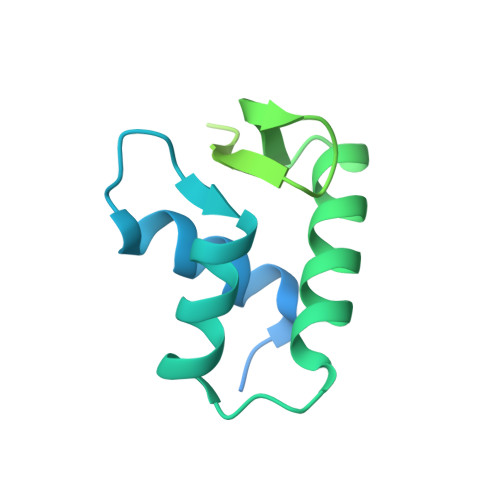
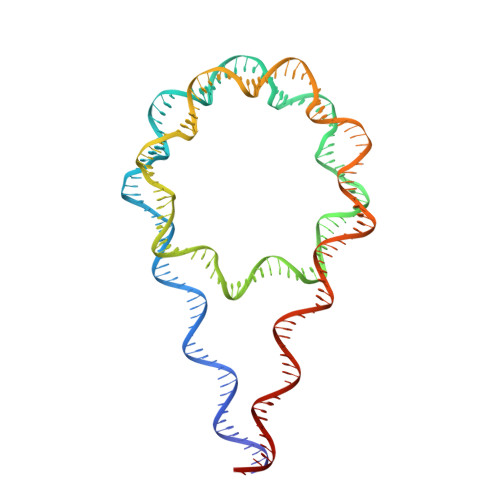
Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1.
Bednar, J., Garcia-Saez, I., Boopathi, R., Cutter, A.R., Papai, G., Reymer, A., Syed, S.H., Lone, I.N., Tonchev, O., Crucifix, C., Menoni, H., Papin, C., Skoufias, D.A., Kurumizaka, H., Lavery, R., Hamiche, A., Hayes, J.J., Schultz, P., Angelov, D., Petosa, C., Dimitrov, S.(2017) Mol Cell 66: 384-397.e8
- PubMed: 28475873
- DOI: https://doi.org/10.1016/j.molcel.2017.04.012
- Primary Citation of Related Structures:
5NL0 - PubMed Abstract:
Linker histones associate with nucleosomes to promote the formation of higher-order chromatin structure, but the underlying molecular details are unclear. We investigated the structure of a 197 bp nucleosome bearing symmetric 25 bp linker DNA arms in complex with vertebrate linker histone H1. We determined electron cryo-microscopy (cryo-EM) and crystal structures of unbound and H1-bound nucleosomes and validated these structures by site-directed protein cross-linking and hydroxyl radical footprinting experiments. Histone H1 shifts the conformational landscape of the nucleosome by drawing the two linkers together and reducing their flexibility. The H1 C-terminal domain (CTD) localizes primarily to a single linker, while the H1 globular domain contacts the nucleosome dyad and both linkers, associating more closely with the CTD-distal linker. These findings reveal that H1 imparts a strong degree of asymmetry to the nucleosome, which is likely to influence the assembly and architecture of higher-order structures.
Organizational Affiliation:
Institut for Advanced Biosciences, Inserm U 1209, CNRS UMR 5309, Université Grenoble Alpes, 38000 Grenoble, France.