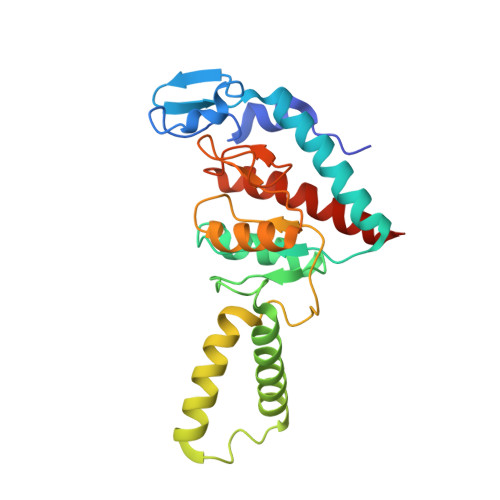
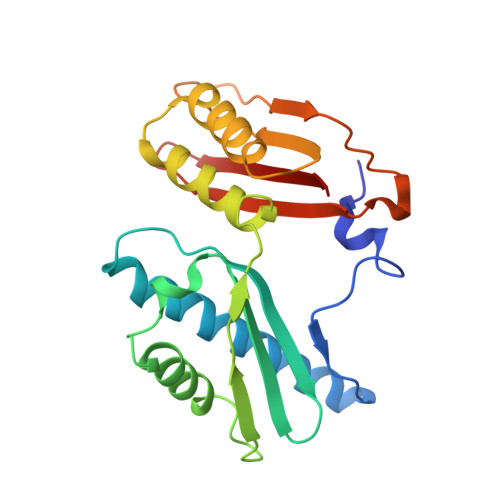
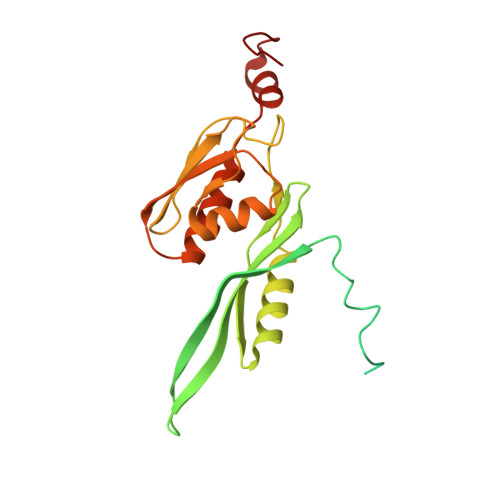
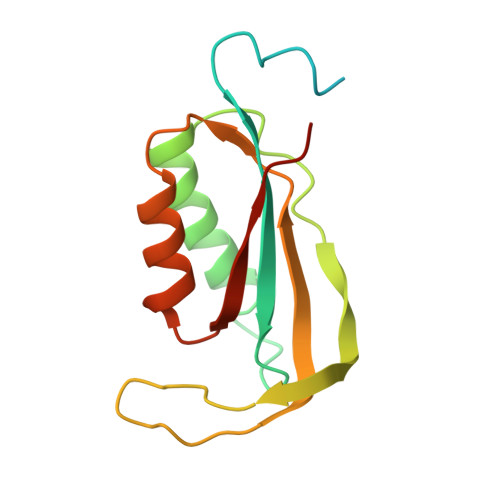
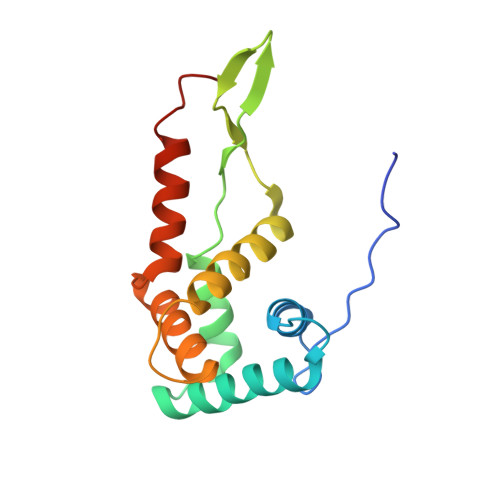
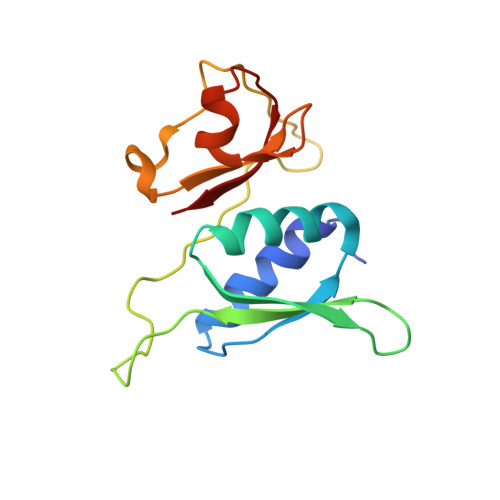
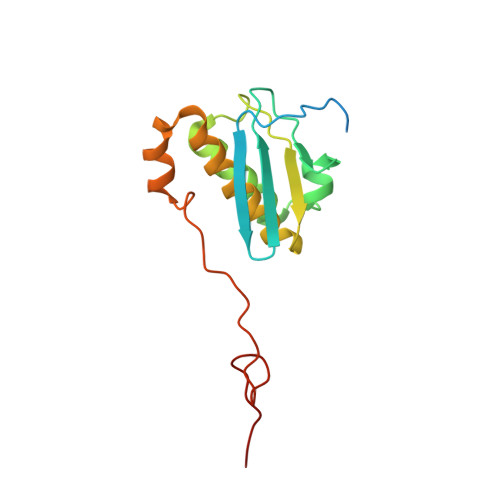
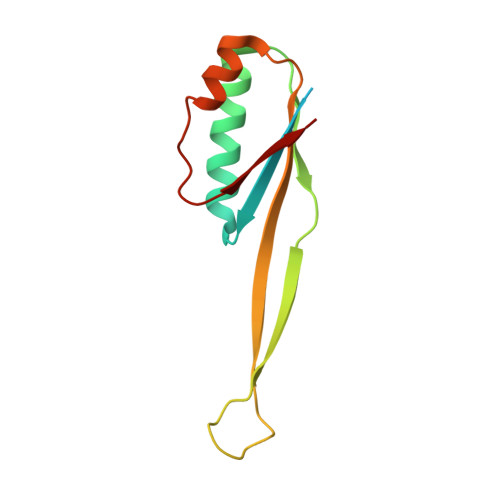
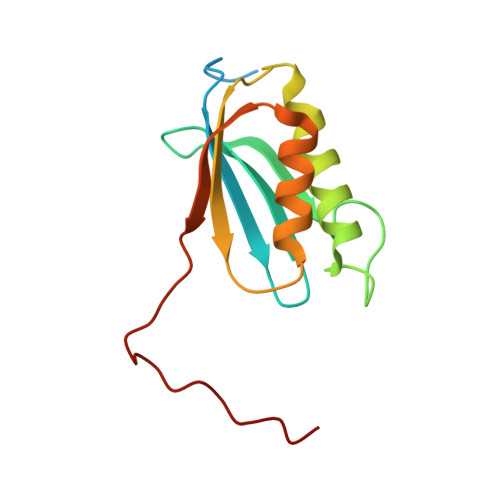
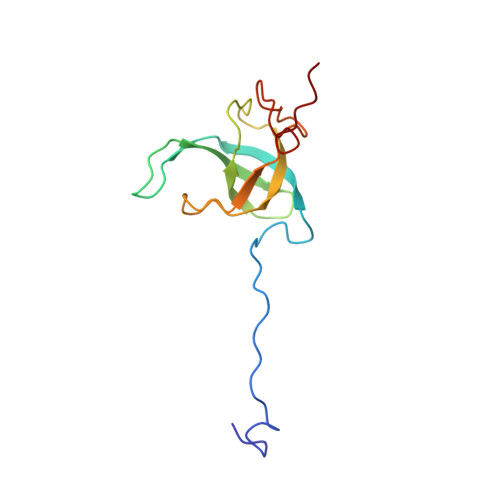
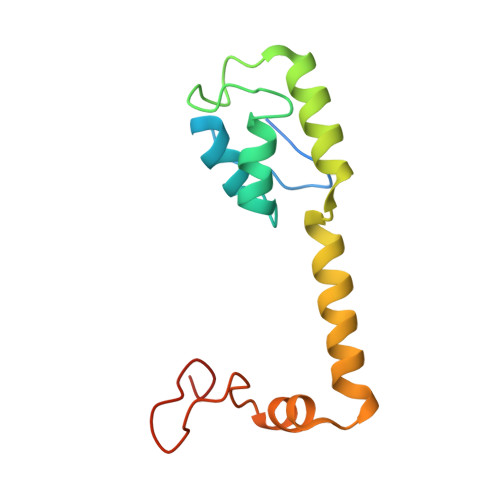
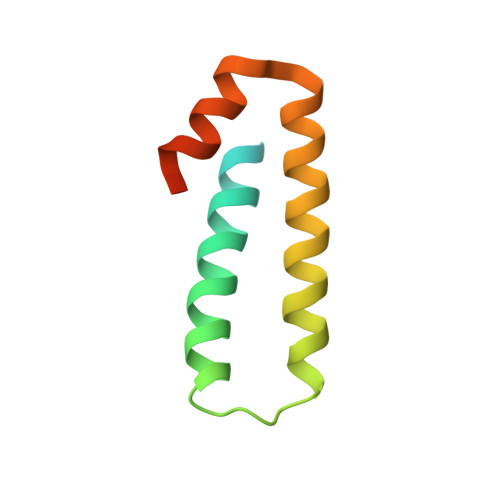
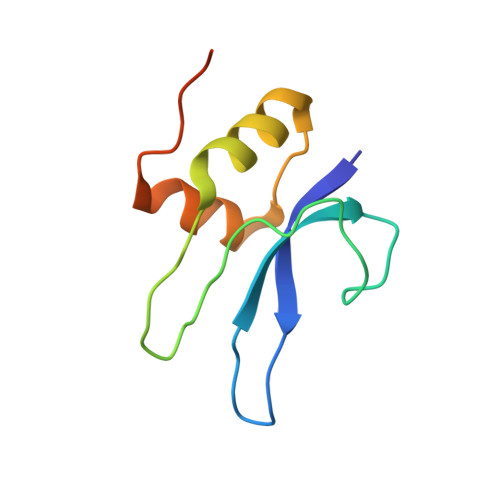
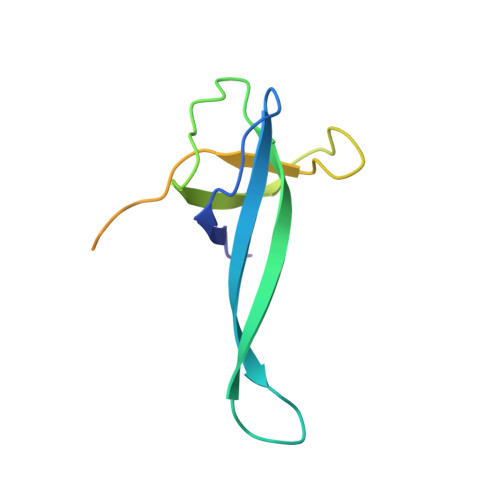
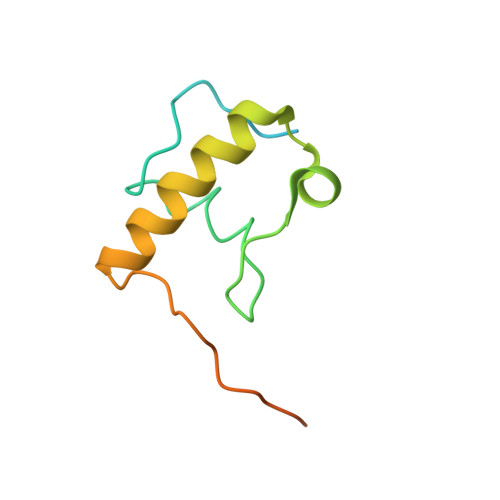
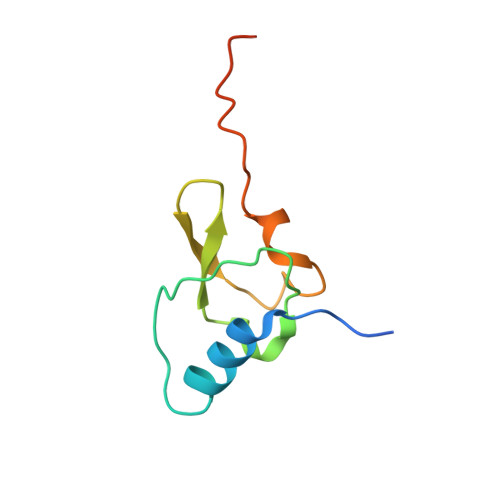
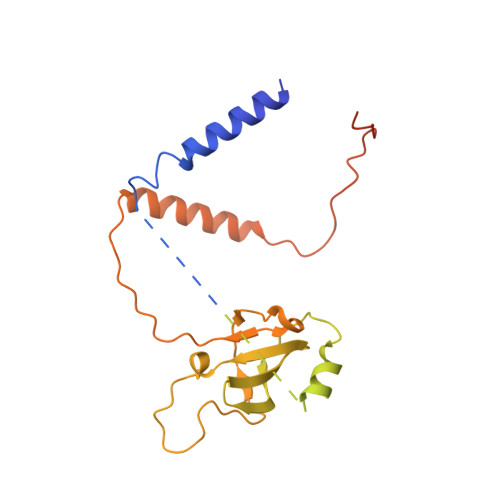
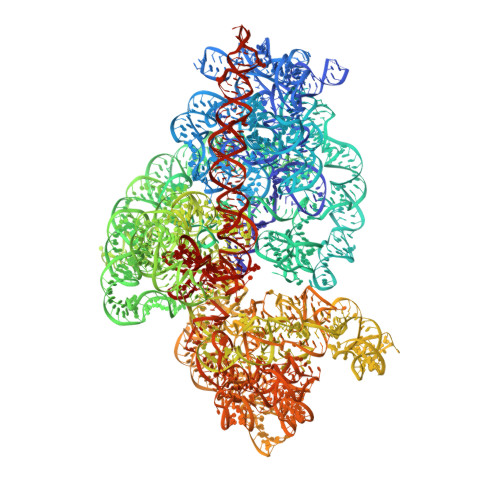
Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Ahmed, T., Shi, J., Bhushan, S.(2017) Nucleic Acids Res 45: 8581-8595
- PubMed: 28582576
- DOI: https://doi.org/10.1093/nar/gkx499
- Primary Citation of Related Structures:
5X8P, 5X8R, 5X8T - PubMed Abstract:
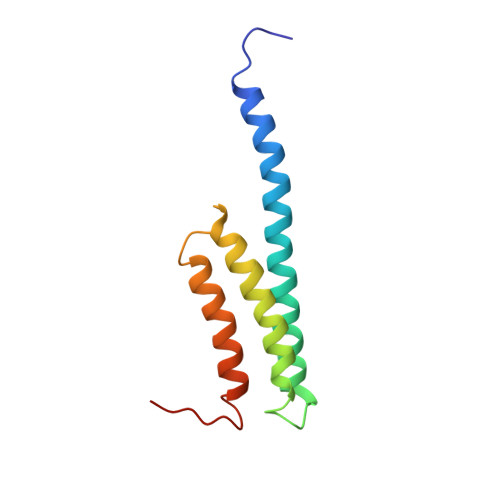
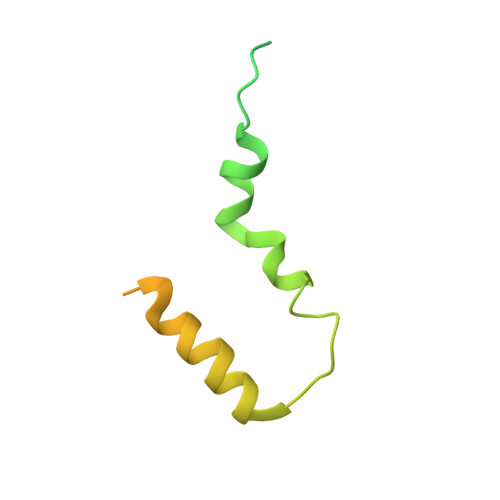
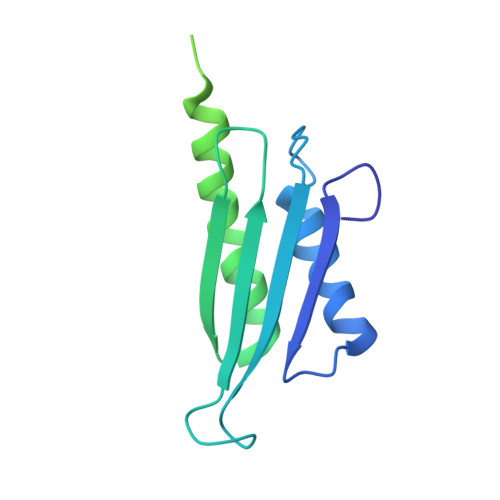
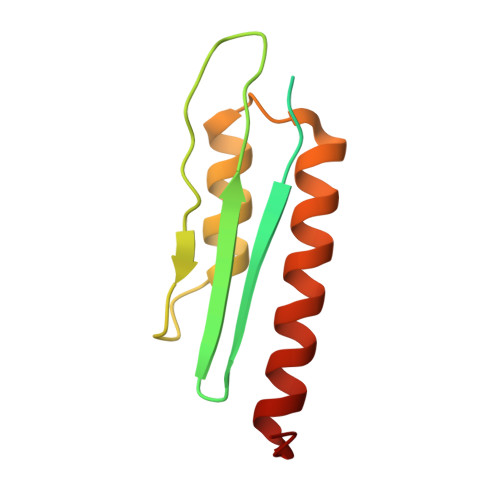
Chloroplastic translation is mediated by a bacterial-type 70S chloroplast ribosome. During the evolution, chloroplast ribosomes have acquired five plastid-specific ribosomal proteins or PSRPs (cS22, cS23, bTHXc, cL37 and cL38) which have been suggested to play important regulatory roles in translation. However, their exact locations on the chloroplast ribosome remain elusive due to lack of a high-resolution structure, hindering our progress to understand their possible roles. Here we present a cryo-EM structure of the 70S chloroplast ribosome from spinach resolved to 3.4 Å and focus our discussion mainly on the architecture of the 30S small subunit (SSU) which is resolved to 3.7 Å. cS22 localizes at the SSU foot where it seems to compensate for the deletions in 16S rRNA. The mRNA exit site is highly remodeled due to the presence of cS23 suggesting an alternative mode of translation initiation. bTHXc is positioned at the SSU head and appears to stabilize the intersubunit bridge B1b during thermal fluctuations. The translation factor plastid pY binds to the SSU on the intersubunit side and interacts with the conserved nucleotide bases involved in decoding. Most of the intersubunit bridges are conserved compared to the bacteria, except for a new bridge involving uL2c and bS6c.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 637551, Singapore.