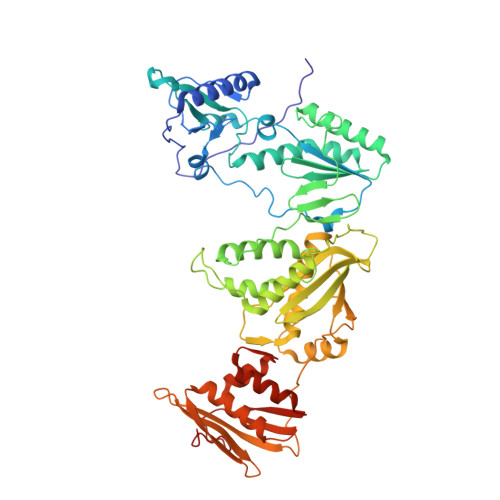
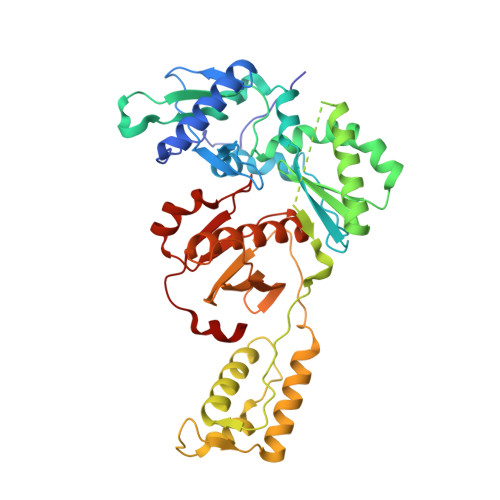
Molecular and cellular studies evaluating a potent 2-cyanoindolizine catechol diether NNRTI targeting wildtype and Y181C mutant HIV-1 reverse transcriptase.
Sasaki, T., Gannam, Z.T.K., Kudalkar, S.N., Frey, K.M., Lee, W.G., Spasov, K.A., Jorgensen, W.L., Anderson, K.S.(2019) Bioorg Med Chem Lett 29: 2182-2188
- PubMed: 31281023
- DOI: https://doi.org/10.1016/j.bmcl.2019.06.047
- Primary Citation of Related Structures:
6DTW, 6DTX - PubMed Abstract:
The development of efficacious NNRTIs for HIV/AIDS therapy is commonly met with the emergence of drug resistant strains, including the Y181C variant. Using a computationally-guided approach, we synthesized the catechol diether series of NNRTIs, which display sub-nanomolar potency in cellular assays. Among the most potent were a series of 2-cyanoindolizine substituted catechol diethers, including Compound 1. We present here a thorough evaluation of this compound, including biochemical, cellular, and structural studies. The compound demonstrates low nanomolar potency against both WT and Y181C HIV-1 RT in in vitro and cellular assays. Our crystal structures of both the wildtype and mutant forms of RT in complex with Compound 1 allow the interrogation of this compound's features that allow it to maintain strong efficacy against the drug resistant mutant. Among these are compensatory shifts in the NNRTI binding pocket, persistence of multiple hydrogen bonds, and van der Waals contacts throughout the binding site. Further, the fluorine at the C6 position of the indolizine moiety makes multiple favorable interactions with both RT forms. The present study highlights the indolizine-substituted catechol diether class of NNRTIs as promising therapeutic candidates possessing optimal pharmacological properties and significant potency against multiple RT variants.
Organizational Affiliation:
Department of Pharmacology, Yale University, 333 Cedar Street, New Haven, CT 06520, United States.