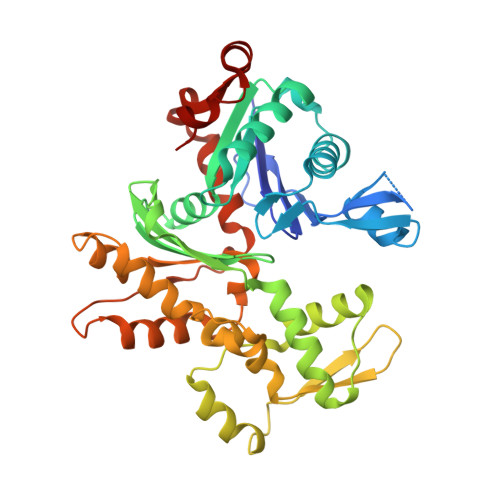
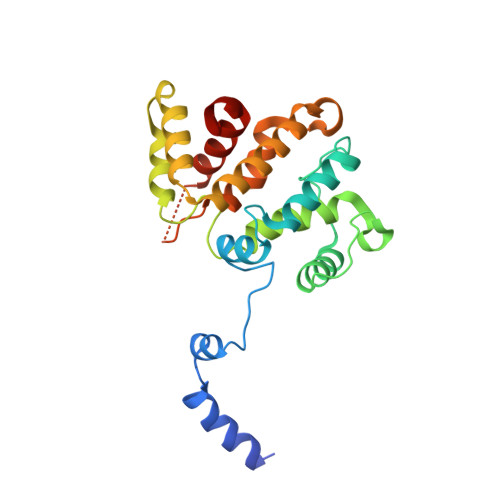
RPEL-family rhoGAPs link Rac/Cdc42 GTP loading to G-actin availability.
Diring, J., Mouilleron, S., McDonald, N.Q., Treisman, R.(2019) Nat Cell Biol 21: 845-855
- PubMed: 31209295
- DOI: https://doi.org/10.1038/s41556-019-0337-y
- Primary Citation of Related Structures:
6GVC - PubMed Abstract:
RPEL proteins, which contain the G-actin-binding RPEL motif, coordinate cytoskeletal processes with actin dynamics. We show that the ArhGAP12- and ArhGAP32-family GTPase-activating proteins (GAPs) are RPEL proteins. We determine the structure of the ArhGAP12/G-actin complex, and show that G-actin contacts the RPEL motif and GAP domain sequences. G-actin inhibits ArhGAP12 GAP activity, and this requires the G-actin contacts identified in the structure. In B16 melanoma cells, ArhGAP12 suppresses basal Rac and Cdc42 activity, F-actin assembly, invadopodia formation and experimental metastasis. In this setting, ArhGAP12 mutants defective for G-actin binding exhibit more effective downregulation of Rac GTP loading following HGF stimulation and enhanced inhibition of Rac-dependent processes, including invadopodia formation. Potentiation or disruption of the G-actin/ArhGAP12 interaction, by treatment with the actin-binding drugs latrunculin B or cytochalasin D, has corresponding effects on Rac GTP loading. The interaction of G-actin with RPEL-family rhoGAPs thus provides a negative feedback loop that couples Rac activity to actin dynamics.
Organizational Affiliation:
Signalling and Transcription Group, The Francis Crick Institute, London, UK.