Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Sun, G., Hu, C., Mei, Q., Luo, M., Chen, X., Li, Z., Liu, Y., Deng, Z., Zhang, Z., Sun, Y.(2020) Nat Commun 11: 4501-4501
- PubMed: 32908132
- DOI: https://doi.org/10.1038/s41467-020-18336-5
- Primary Citation of Related Structures:
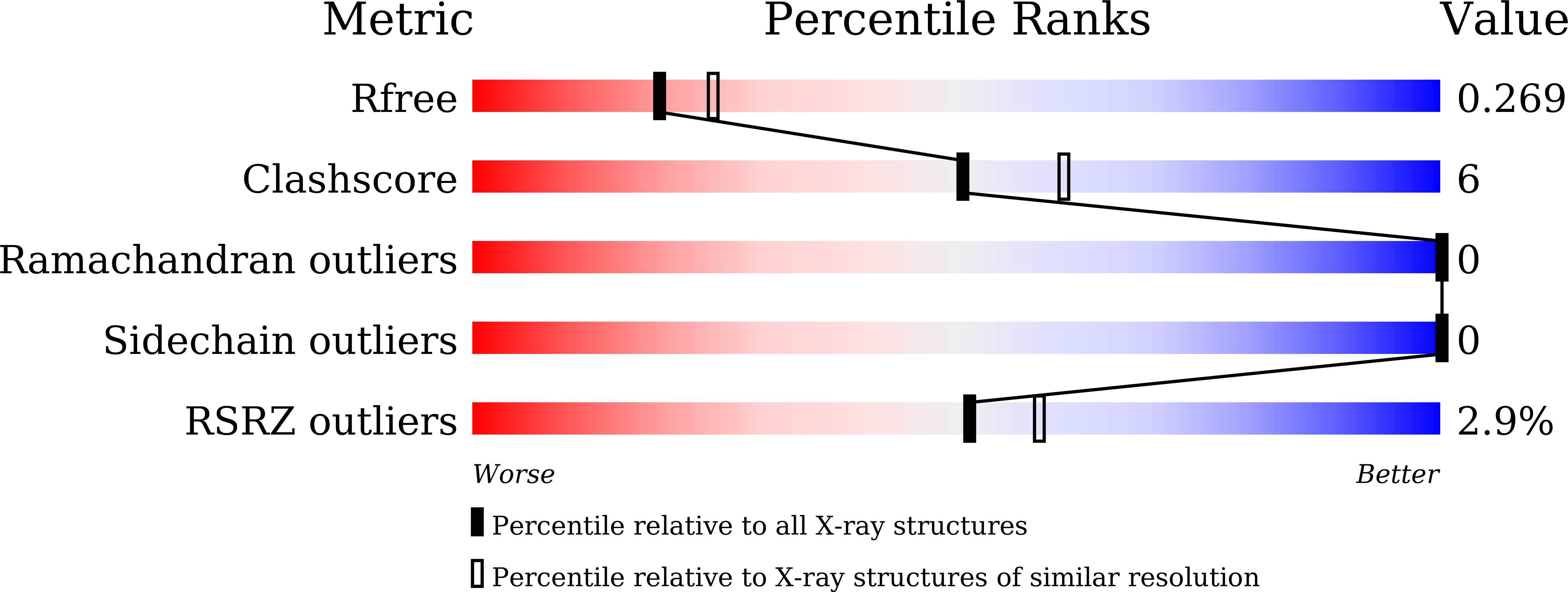
6M4P, 6M4Q - PubMed Abstract:
Streptovaricin C is a naphthalenic ansamycin antibiotic structurally similar to rifamycins with potential anti-MRSA bioactivities. However, the formation mechanism of the most fascinating and bioactivity-related methylenedioxy bridge (MDB) moiety in streptovaricins is unclear. Based on genetic and biochemical evidences, we herein clarify that the P450 enzyme StvP2 catalyzes the MDB formation in streptovaricins, with an atypical substrate inhibition kinetics. Furthermore, X-ray crystal structures in complex with substrate and structure-based mutagenesis reveal the intrinsic details of the enzymatic reaction. The mechanism of MDB formation is proposed to be an intramolecular nucleophilic substitution resulting from the hydroxylation by the heme core and the keto-enol tautomerization via a crucial catalytic triad (Asp89-His92-Arg72) in StvP2. In addition, in vitro reconstitution uncovers that C6-O-methylation and C4-O-acetylation of streptovaricins are necessary prerequisites for the MDB formation. This work provides insight for the MDB formation and adds evidence in support of the functional versatility of P450 enzymes.
Organizational Affiliation:
Key Laboratory of Combinatorial Biosynthesis and Drug Discovery, Ministry of Education, Wuhan University School of Pharmaceutical Sciences, 430071, Wuhan, P. R. China.