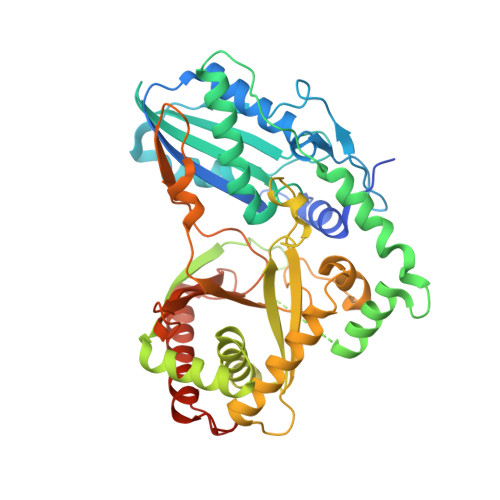
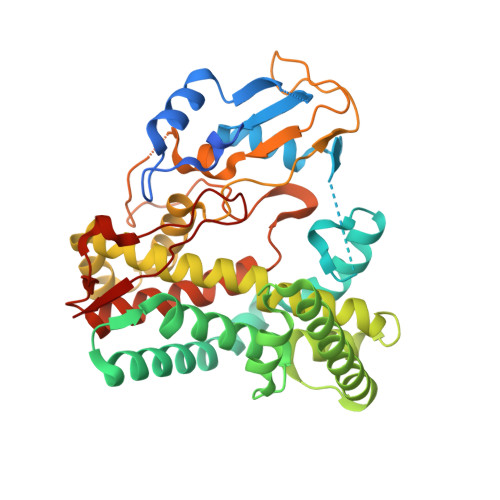
Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics.
Greule, A., Izore, T., Iftime, D., Tailhades, J., Schoppet, M., Zhao, Y., Peschke, M., Ahmed, I., Kulik, A., Adamek, M., Goode, R.J.A., Schittenhelm, R.B., Kaczmarski, J.A., Jackson, C.J., Ziemert, N., Krenske, E.H., De Voss, J.J., Stegmann, E., Cryle, M.J.(2019) Nat Commun 10: 2613-2613
- PubMed: 31197182
- DOI: https://doi.org/10.1038/s41467-019-10384-w
- Primary Citation of Related Structures:
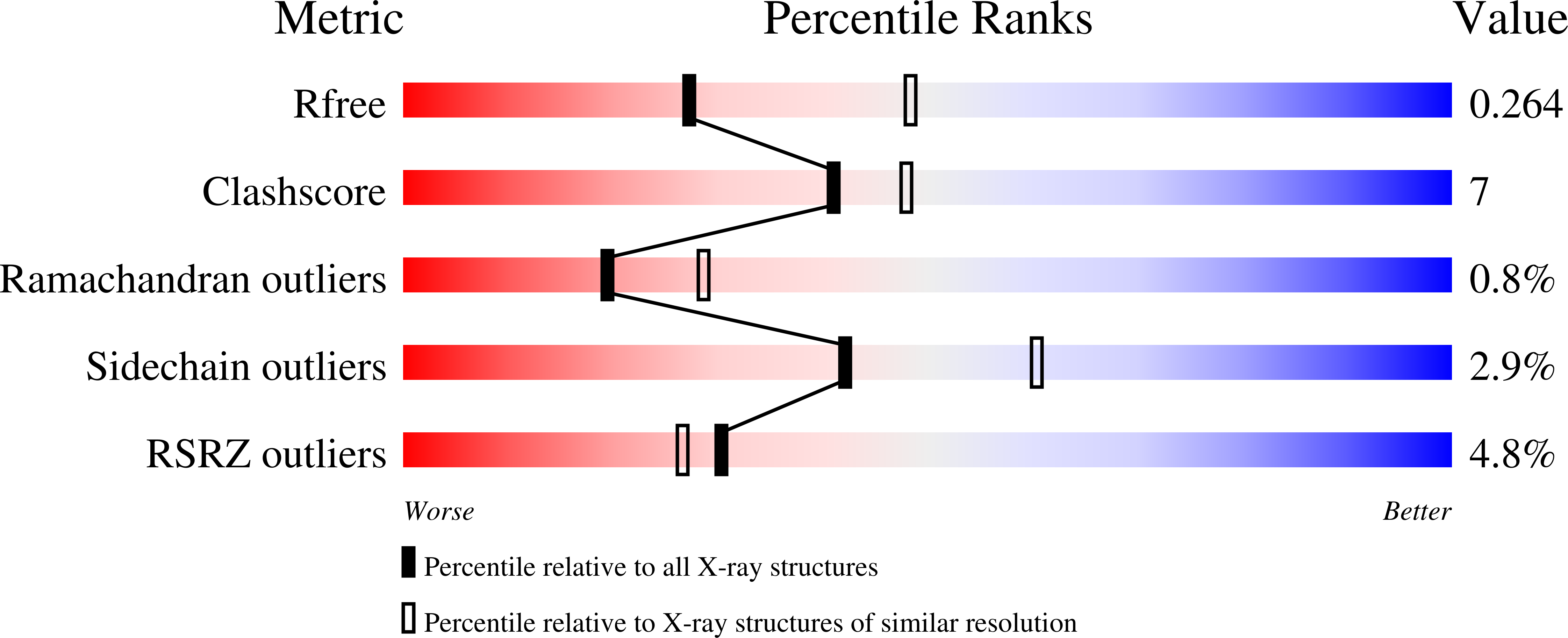
6M7L - PubMed Abstract:
Kistamicin is a divergent member of the glycopeptide antibiotics, a structurally complex class of important, clinically relevant antibiotics often used as the last resort against resistant bacteria. The extensively crosslinked structure of these antibiotics that is essential for their activity makes their chemical synthesis highly challenging and limits their production to bacterial fermentation. Kistamicin contains three crosslinks, including an unusual 15-membered A-O-B ring, despite the presence of only two Cytochrome P450 Oxy enzymes thought to catalyse formation of such crosslinks within the biosynthetic gene cluster. In this study, we characterise the kistamicin cyclisation pathway, showing that the two Oxy enzymes are responsible for these crosslinks within kistamicin and that they function through interactions with the X-domain, unique to glycopeptide antibiotic biosynthesis. We also show that the kistamicin OxyC enzyme is a promiscuous biocatalyst, able to install multiple crosslinks into peptides containing phenolic amino acids.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The Monash Biomedicine Discovery Institute, Monash University, Clayton, VIC, 3800, Australia.