Structural Refinement of the Tubulin Ligand (+)-Discodermolide to Attenuate Chemotherapy-Mediated Senescence.
Guo, B., Rodriguez-Gabin, A., Prota, A.E., Muhlethaler, T., Zhang, N., Ye, K., Steinmetz, M.O., Horwitz, S.B., Smith 3rd, A.B., McDaid, H.M.(2020) Mol Pharmacol 98: 156-167
- PubMed: 32591477
- DOI: https://doi.org/10.1124/mol.119.117457
- Primary Citation of Related Structures:
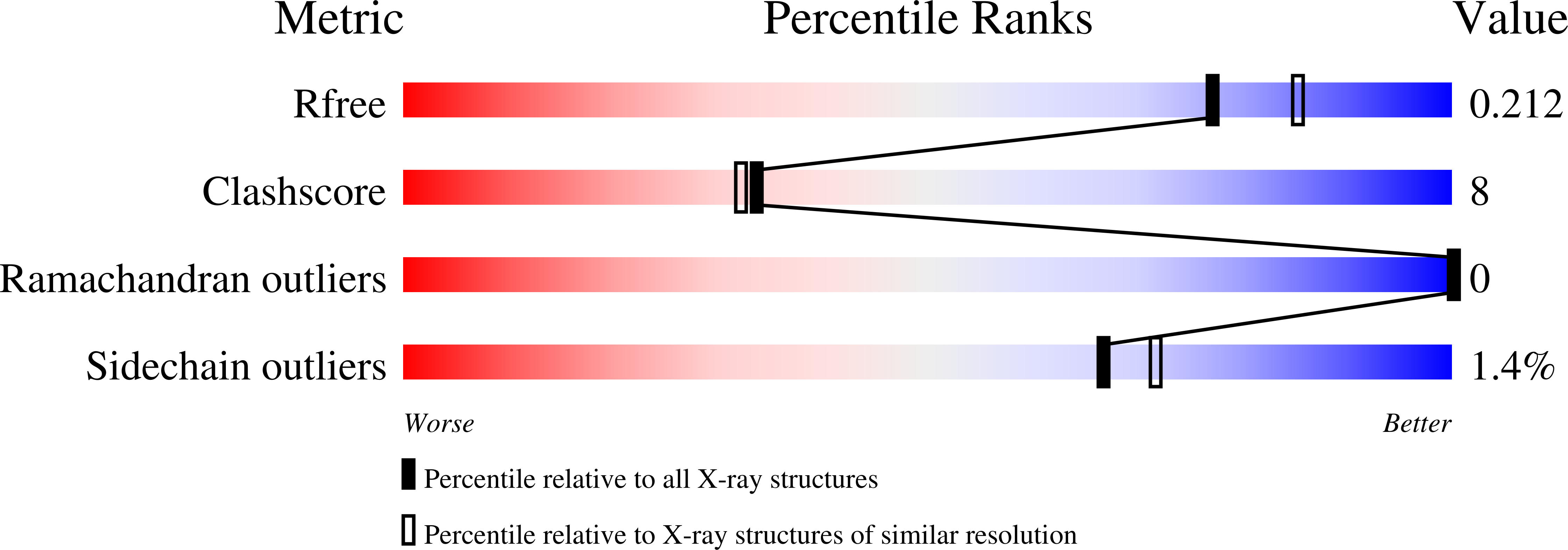
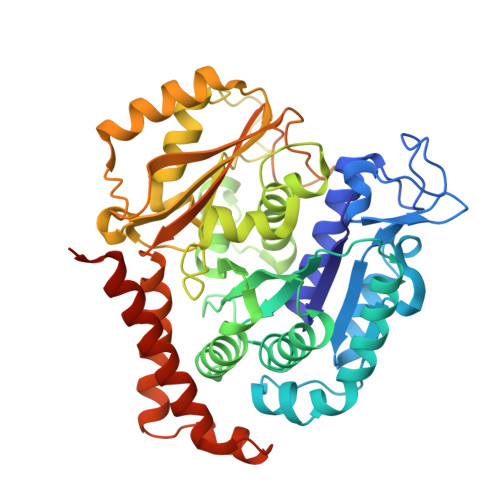
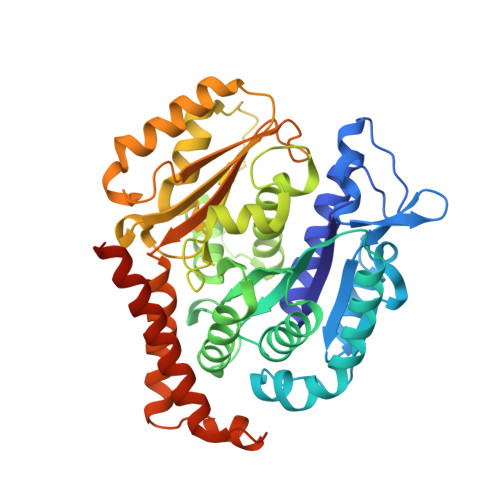
6SES - PubMed Abstract:
The natural product (+)-discodermolide (DDM) is a microtubule stabilizing agent and potent inducer of senescence. We refined the structure of DDM and evaluated the activity of novel congeners in triple negative breast and ovarian cancers, malignancies that typically succumb to taxane resistance. Previous structure-activity analyses identified the lactone and diene as moieties conferring anticancer activity, thus identifying priorities for the structural refinement studies described herein. Congeners possessing the monodiene with a simplified lactone had superior anticancer efficacy relative to taxol, particularly in resistant models. Specifically, one of these congeners, B2, demonstrated 1) improved pharmacologic properties, specifically increased maximum response achievable and area under the curve, and decreased EC 50 ; 2) a uniform dose-response profile across genetically heterogeneous cancer cell lines relative to taxol or DDM; 3) reduced propensity for senescence induction relative to DDM; 4) superior long-term activity in cancer cells versus taxol or DDM; and 5) attenuation of metastatic characteristics in treated cancer cells. To contrast the binding of B2 versus DDM in tubulin, X-ray crystallography studies revealed a shift in the position of the lactone ring associated with removal of the C2-methyl and C3-hydroxyl. Thus, B2 may be more adaptable to changes in the taxane site relative to DDM that could account for its favorable properties. In conclusion, we have identified a DDM congener with broad range anticancer efficacy that also has decreased risk of inducing chemotherapy-mediated senescence. SIGNIFICANCE STATEMENT: Here, we describe the anticancer activity of novel congeners of the tubulin-polymerizing molecule (+)-discodermolide. A lead molecule is identified that exhibits an improved dose-response profile in taxane-sensitive and taxane-resistant cancer cell models, diminished risk of chemotherapy-mediated senescence, and suppression of tumor cell invasion endpoints. X-ray crystallography studies identify subtle changes in the pose of binding to β -tubulin that could account for the improved anticancer activity. These findings support continued preclinical development of discodermolide, particularly in the chemorefractory setting.
Organizational Affiliation:
Department of Chemistry, Monell Chemical Senses Center and Laboratory for Research on the Structure of Matter, University of Pennsylvania, Philadelphia, Pennsylvania (B.G., N.Z., A.B.S.); Departments of Molecular Pharmacology (A.R.-G., S.B.H., H.M.M.), Epidemiology (K.Y.), and Medicine (H.M.M.), Albert Einstein College of Medicine, Bronx, New York; Laboratory of Biomolecular Research, Division of Biology and Chemistry, Paul Scherrer Institut, Villigen, Switzerland (A.E.P., T.M., M.O.S.); and University of Basel, Biozentrum, Basel, Switzerland (M.O.S.).