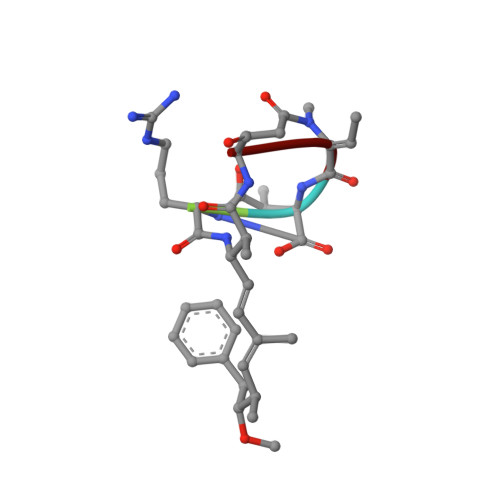
Solution structure of nodularin. An inhibitor of serine/threonine-specific protein phosphatases.
Annila, A., Lehtimaki, J., Mattila, K., Eriksson, J.E., Sivonen, K., Rantala, T.T., Drakenberg, T.(1996) J Biol Chem 271: 16695-16702
- PubMed: 8663277
- DOI: https://doi.org/10.1074/jbc.271.28.16695
- Primary Citation of Related Structures:
1AY3 - PubMed Abstract:
The three-dimensional solution structure of nodularin was studied by NMR and molecular dynamics simulations. The conformation in water was determined from the distance and dihedral data by distance geometry and refined by iterative relaxation matrix analysis. The cyclic backbone adopts a well defined conformation but the remote parts of the side chains of arginine as well as the amino acid derivative Adda have a large spatial dispersion. For the unusual amino acids the partial charges were calculated and nodularin was subjected to molecular dynamic simulations in water. A good agreement was found between experimental and computational data with hydrogen bonds, solvent accessibility, molecular motion, and conformational exchange. The three-dimensional structure resembles very closely that of microcystin-LR in the chemically equivalent segment. Therefore, it is expected that the binding of both microcystins and nodularins to serine/threonine-specific protein phosphatases is similar on an atomic level.
Organizational Affiliation:
VTT Chemical Technology, P. O. Box 1401, FIN-02044 VTT, Finland.