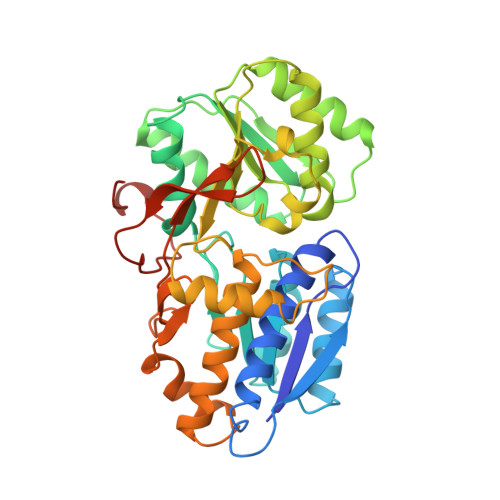
Crystal structure of AmiC: the controller of transcription antitermination in the amidase operon of Pseudomonas aeruginosa.
Pearl, L., O'Hara, B., Drew, R., Wilson, S.(1994) EMBO J 13: 5810-5817
- PubMed: 7813419
- DOI: https://doi.org/10.1002/j.1460-2075.1994.tb06924.x
- Primary Citation of Related Structures:
1PEA - PubMed Abstract:
The crystal structure for the negative regulator (AmiC) of the amidase operon from Pseudomonas aeruginosa has been solved at a resolution of 2.1 A. AmiC is the amide sensor protein in the amidase operon and regulates the activity of the transcription antitermination factor AmiR, which in turn regulates amidase expression. The AmiC structure consists of two domains with an alternating beta-alpha-beta topology. The two domains are separated by a central cleft and the amide binding site is positioned in this cleft at the interface of the domains. The overall fold for AmiC is extremely similar to that for the leucine-isoleucine-valine binding protein (LivJ) of Escherichia coli despite only 17% sequence identity, however, the two domains of AmiC are substantially closed compared with LivJ. The closed structure of AmiC is stabilized significantly by the bound acetamide, suggesting a molecular mechanism for the process of amide induction. The amide binding site is extremely specific for acetamide and would not allow a closed conformation in the presence of the anti-inducer molecule butyramide.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University College London, UK.