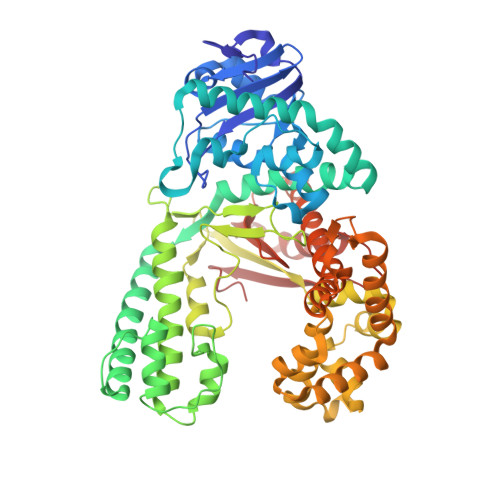
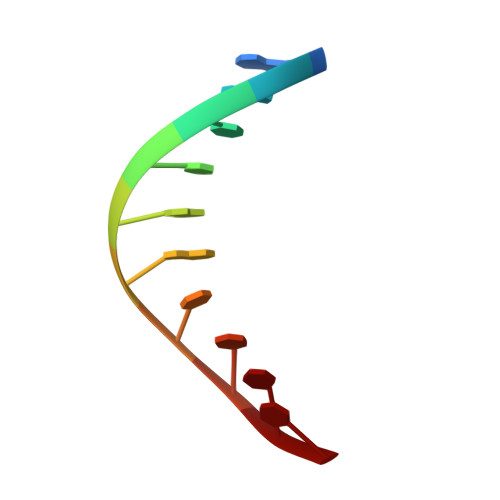
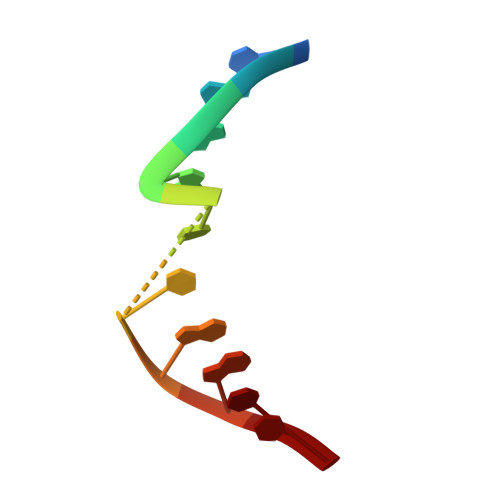
Crystal Structures and Repair Studies Reveal the Identity and the Base-Pairing Properties of the Uv-Induced Spore Photoproduct DNA Lesion.
Heil, K., Kneuttinger, A.C., Schneider, S., Lischke, U., Carell, T.(2011) Chemistry 17: 9651
- PubMed: 21780197
- DOI: https://doi.org/10.1002/chem.201100177
- Primary Citation of Related Structures:
2Y1I, 2Y1J - PubMed Abstract:
UV light is one of the major causes of DNA damage. In spore DNA, due to an unusual packing of the genetic material, a special spore photoproduct lesion (SP lesion) is formed, which is repaired by the enzyme spore photoproduct lyase (Spl), a radical S-adenosylmethionine (SAM) enzyme. We report here the synthesis and DNA incorporation of a DNA SP lesion analogue lacking the phosphodiester backbone. The oligonucleotides were used for repair studies and they were cocrystallized with a polymerase enzyme as a template to clarify the configuration of the SP lesion and to provide information about the base-pairing properties of the lesion. The structural analysis together with repair studies allowed us to clarify the identity of the preferentially repaired lesion diastereoisomer.
Organizational Affiliation:
Center for Integrated Protein Science (CiPSM) at the Department of Chemistry, Ludwig-Maximilians-University, Munich, Germany.