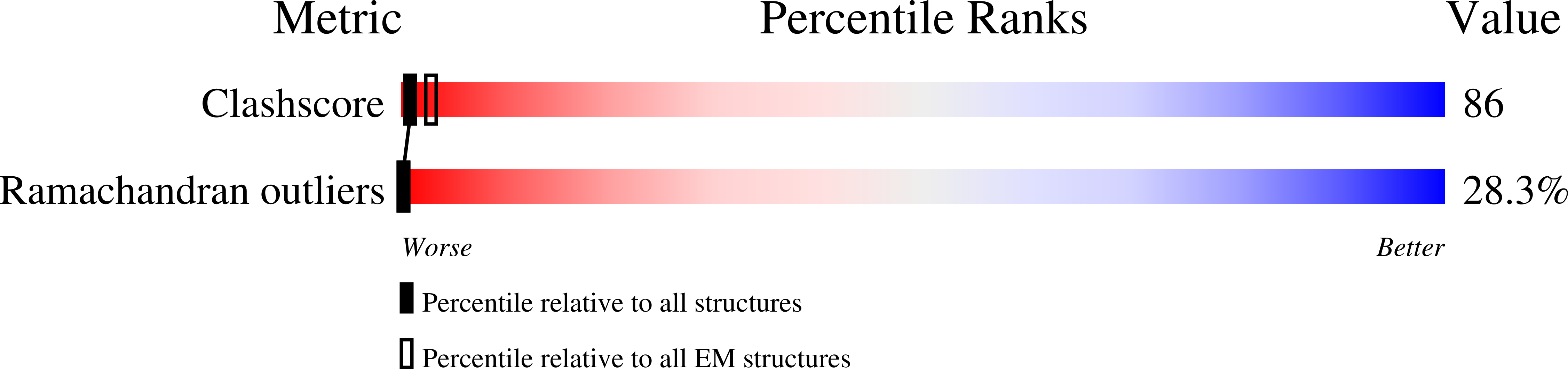Structural basis for TetM-mediated tetracycline resistance.
Donhofer, A., Franckenberg, S., Wickles, S., Berninghausen, O., Beckmann, R., Wilson, D.N.(2012) Proc Natl Acad Sci U S A 109: 16900-16905
- PubMed: 23027944
- DOI: https://doi.org/10.1073/pnas.1208037109
- Primary Citation of Related Structures:
3J25 - PubMed Abstract:
Ribosome protection proteins (RPPs) confer tetracycline resistance by binding to the ribosome and chasing the drug from its binding site. The current model for the mechanism of action of RPPs proposes that drug release is indirect and achieved via conformational changes within the drug-binding site induced upon binding of the RPP to the ribosome. Here we report a cryo-EM structure of the RPP TetM in complex with the 70S ribosome at 7.2-Å resolution. The structure reveals the contacts of TetM with the ribosome, including interaction between the conserved and functionally critical C-terminal extension of TetM and the decoding center of the small subunit. Moreover, we observe direct interaction between domain IV of TetM and the tetracycline binding site and identify residues critical for conferring tetracycline resistance. A model is presented whereby TetM directly dislodges tetracycline to confer resistance.
Organizational Affiliation:
Gene Center and Department for Biochemistry, University of Munich, 81377 Munich, Germany.