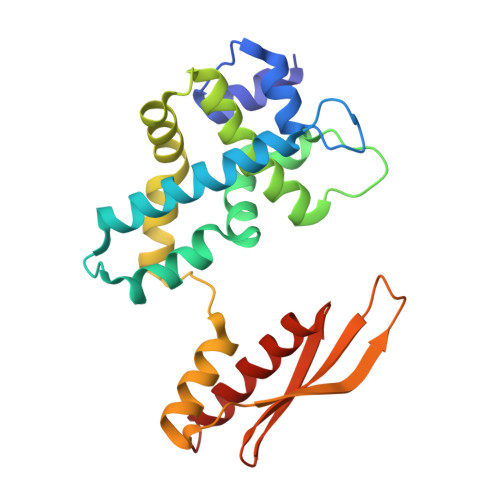
RNase III: Genetics and Function; Structure and Mechanism.
Court, D.L., Gan, J., Liang, Y.H., Shaw, G.X., Tropea, J.E., Costantino, N., Waugh, D.S., Ji, X.(2013) Annu Rev Genet 47: 405-431
- PubMed: 24274754
- DOI: https://doi.org/10.1146/annurev-genet-110711-155618
- Primary Citation of Related Structures:
4M2Z, 4M30 - PubMed Abstract:
RNase III is a global regulator of gene expression in Escherichia coli that is instrumental in the maturation of ribosomal and other structural RNAs. We examine here how RNase III itself is regulated in response to growth and other environmental changes encountered by the cell and how, by binding or processing double-stranded RNA (dsRNA) intermediates, RNase III controls the expression of genes. Recent insight into the mechanism of dsRNA binding and processing, gained from structural studies of RNase III, is reviewed. Structural studies also reveal new cleavage sites in the enzyme that can generate longer 3' overhangs.
Organizational Affiliation:
Center for Cancer Research, National Cancer Institute, Frederick, Maryland 21702; email: courtd@mail.nih.gov , ganj@mail.nih.gov , liangyh@mail.nih.gov , shawg@mail.nih.gov , costantn@mail.nih.gov , tropeajo@mail.nih.gov , waughd@mail.nih.gov , jix@mail.nih.gov.