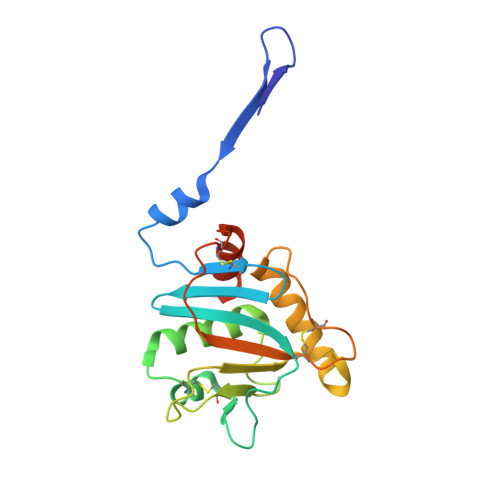
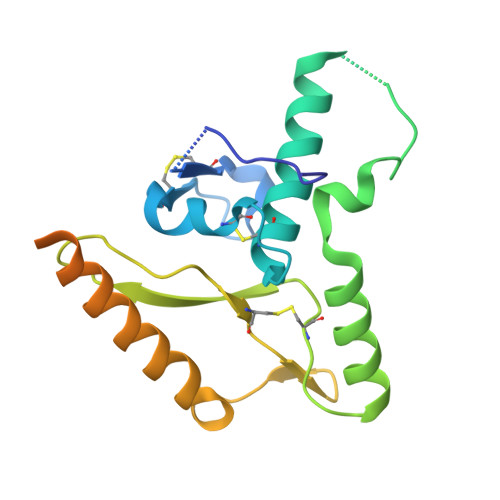
Structure and stabilization of the antigenic glycoprotein building blocks of the New World mammarenavirus spike complex.
Paesen, G.C., Ng, W.M., Kimuda, S., Sutton, G., Doores, K.J., Bowden, T.A.(2025) mBio 16: e0107625-e0107625
- PubMed: 40511941
- DOI: https://doi.org/10.1128/mbio.01076-25
- Primary Citation of Related Structures:
9GHI, 9GHJ, 9QQN - PubMed Abstract:
The spillover of New World (NW) arenaviruses from rodent reservoirs into human populations poses a continued risk to human health. NW arenaviruses present a glycoprotein (GP) complex on the envelope surface of the virion, which orchestrates host cell entry and is a key target of the immune response arising from infection and immunization. Each protomer of the trimeric GP is composed of a stable signal peptide, a GP1 attachment glycoprotein, and a GP2 fusion glycoprotein. To glean insights into the architecture of this key therapeutic target, we determined the crystal structures of NW GP1-GP2 heterodimeric complexes from Junín virus and Machupo virus. Due to the metastability of the interaction between GP1 and GP2, structural elucidation required the introduction of a disulfide bond at the GP1-GP2 complex interface, but no other stabilizing modifications were required. While the overall assembly of NW GP1-GP2 is conserved with that presented by Old World (OW) arenaviruses, including Lassa virus and lymphocytic choriomeningitis virus, NW GP1-GP2 complexes are structurally distinct. Indeed, we note that when compared to the OW GP1-GP2 complex, the globular portion of NW GP1 undergoes limited structural alterations upon detachment from its cognate GP2. We further demonstrate that our engineered GP1-GP2 heterodimers are antigenically relevant and recognized by neutralizing antibodies. These data provide insights into the distinct assemblies presented by NW and OW arenaviruses, as well as provide molecular-level blueprints that may guide vaccine development.IMPORTANCEAlthough the emergence of New World (NW) hemorrhagic fever mammarenaviruses poses an unceasing threat to human health, there is a paucity of reagents capable of protecting against the transmission of these pathogens from their natural rodent reservoirs. This is, in part, attributed to our limited understanding of the structure and function of the NW glycoprotein spike complex presented on the NW arenavirus surface. Here, we provide a detailed molecular-level description of how the two major components of this key therapeutic target assemble to form a key building block of the NW arenaviral spike complex. The insights gleaned from this work provide a framework for guiding the structure-based development of NW arenaviral vaccines.
Organizational Affiliation:
Division of Structural Biology, Centre for Human Genetics, University of Oxford, Oxford, United Kingdom.