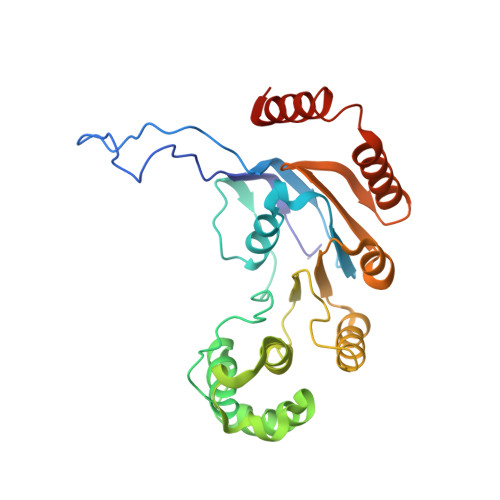
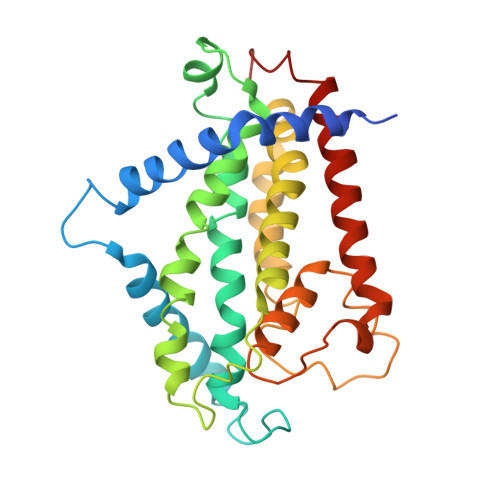
Cryo-electron Microscopy Structure and Transport Mechanism of a Wall Teichoic Acid ABC Transporter.
Chen, L., Hou, W.T., Fan, T., Liu, B., Pan, T., Li, Y.H., Jiang, Y.L., Wen, W., Chen, Z.P., Sun, L., Zhou, C.Z., Chen, Y.(2020) mBio 11
- PubMed: 32184247
- DOI: https://doi.org/10.1128/mBio.02749-19
- Primary Citation of Related Structures:
6JBH - PubMed Abstract:
The wall teichoic acid (WTA) is a major cell wall component of Gram-positive bacteria, such as methicillin-resistant Staphylococcus aureus (MRSA), a common cause of fatal clinical infections in humans. Thus, the indispensable ABC transporter TarGH, which flips WTA from cytoplasm to extracellular space, becomes a promising target of anti-MRSA drugs. Here, we report the 3.9-Å cryo-electron microscopy (cryo-EM) structure of a 50% sequence-identical homolog of TarGH from Alicyclobacillus herbarius at an ATP-free and inward-facing conformation. Structural analysis combined with activity assays enables us to clearly decode the binding site and inhibitory mechanism of the anti-MRSA inhibitor Targocil, which targets TarGH. Moreover, we propose a "crankshaft conrod" mechanism utilized by TarGH, which can be applied to similar ABC transporters that translocate a rather big substrate through relatively subtle conformational changes. These findings provide a structural basis for the rational design and optimization of antibiotics against MRSA. IMPORTANCE The wall teichoic acid (WTA) is a major component of cell wall and a pathogenic factor in methicillin-resistant Staphylococcus aureus (MRSA). The ABC transporter TarGH is indispensable for flipping WTA precursor from cytoplasm to the extracellular space, thus making it a promising drug target for anti-MRSA agents. The 3.9-Å cryo-EM structure of a TarGH homolog helps us to decode the binding site and inhibitory mechanism of a recently reported inhibitor, Targocil, and provides a structural platform for rational design and optimization of potential antibiotics. Moreover, we propose a "crankshaft conrod" mechanism to explain how a big substrate is translocated through subtle conformational changes of type II exporters. These findings advance our understanding of anti-MRSA drug design and ABC transporters.
- Hefei National Laboratory for Physical Sciences at the Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui, China.
Organizational Affiliation: