Autologous Antibody Responses to an HIV Envelope Glycan Hole Are Not Easily Broadened in Rabbits.
Yang, Y.R., McCoy, L.E., van Gils, M.J., Andrabi, R., Turner, H.L., Yuan, M., Cottrell, C.A., Ozorowski, G., Voss, J., Pauthner, M., Polveroni, T.M., Messmer, T., Wilson, I.A., Sanders, R.W., Burton, D.R., Ward, A.B.(2020) J Virol 94
- PubMed: 31941772
- DOI: https://doi.org/10.1128/JVI.01861-19
- Primary Citation of Related Structures:
6U59 - PubMed Abstract:
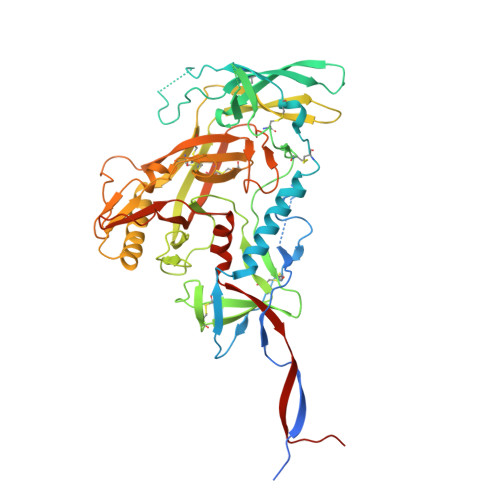
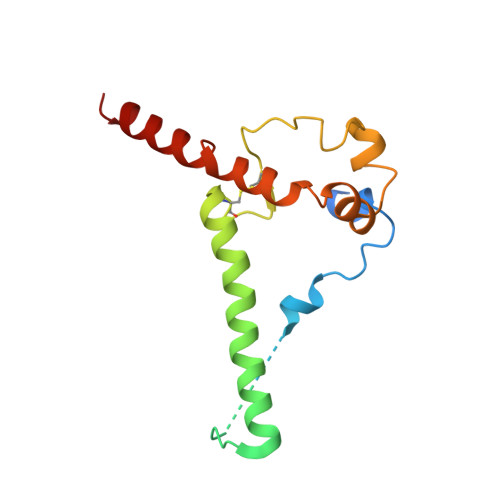
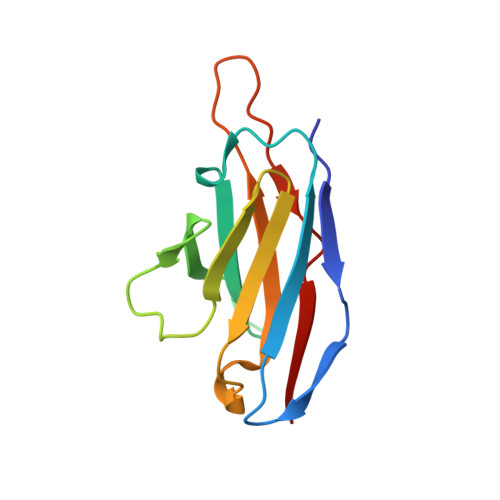
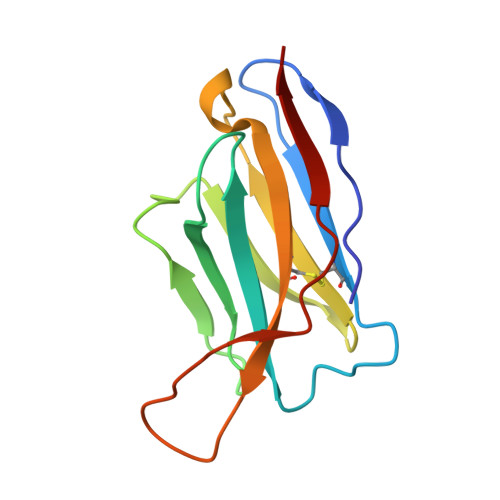
Extensive studies with subtype A BG505-derived HIV envelope glycoprotein (Env) immunogens have revealed that the dominant autologous neutralizing epitope in rabbits is located in an exposed region of the heavily glycosylated trimer that lacks potential N-linked glycosylation sites at positions 230, 241, and 289. The Env derived from B41, a subtype B virus, shares a glycan hole centered on positions 230 and 289. To test whether broader neutralization to the common glycan hole can be achieved, we immunized rabbits with B41 SOSIP (gp120-gp41 disulfide [SOS] with an isoleucine-to-proline mutation [IP] in gp41) alone, as well as B41 and BG505 coimmunization. We isolated autologous neutralizing antibodies (nAbs) and described their structure in complex with the B41 Env. Our data suggest that distinct autologous nAb lineages are induced by BG505 and B41 immunogens, even when both were administered together. In contrast to previously described BG505 glycan hole antibodies, the B41-specific nAbs accommodate the >97% conserved N241 glycan, which is present in B41. Single-particle cryo-electron microscopy studies confirmed that B41- and BG505-specific nAbs bind to overlapping glycan hole epitopes. We then used our high-resolution data to guide mutations in the BG505 glycan hole epitope in an attempt to broaden the reactivity of a B41-specific nAb, but we recovered only partial binding. Our data demonstrate that the lack of cross-reactivity in glycan hole antibodies is due to amino acid differences within the epitope, and our attempts to rationally design cross-reactive trimers resulted in only limited success. Thus, even for the immunodominant glycan hole shared between BG505 and B41, the prospect of designing prime-boost immunogens remains difficult. IMPORTANCE A glycan hole is one of the most dominant autologous neutralizing epitopes targeted on BG505 and B41 SOSIP trimer-immunized rabbits. Our high-resolution cryo-electron microscopy (cryoEM) studies of B41 in complex with a B41-specific antibody complex elucidate the molecular basis of this strain-specific glycan hole response. We conclude that even for the immunodominant glycan hole shared between BG505 and B41, the prospect of designing prime-boost immunogens remains difficult.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, California, USA.