Electron cryomicroscopy observation of acyl carrier protein translocation in type I fungal fatty acid synthase.
Lou, J.W., Iyer, K.R., Hasan, S.M.N., Cowen, L.E., Mazhab-Jafari, M.T.(2019) Sci Rep 9: 12987-12987
- PubMed: 31506493
- DOI: https://doi.org/10.1038/s41598-019-49261-3
- Primary Citation of Related Structures:
6U5T, 6U5U, 6U5V, 6U5W - PubMed Abstract:
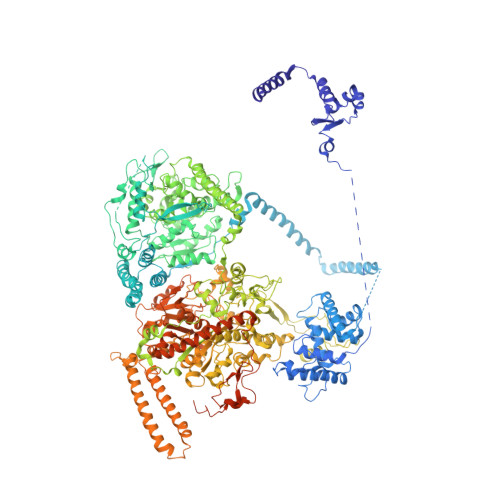
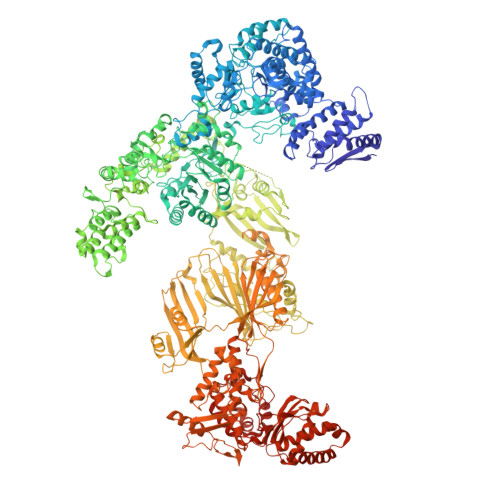
During fatty acid biosynthesis, acyl carrier proteins (ACPs) from type I fungal fatty acid synthase (FAS) shuttle substrates and intermediates within a reaction chamber that hosts multiple spatially-fixed catalytic centers. A major challenge in understanding the mechanism of ACP-mediated substrate shuttling is experimental observation of its transient interaction landscape within the reaction chamber. Here, we have shown that ACP spatial distribution is sensitive to the presence of substrates in a catalytically inhibited state, which enables high-resolution investigation of the ACP-dependent conformational transitions within the enoyl reductase (ER) reaction site. In two fungal FASs with distinct ACP localization, the shuttling domain is targeted to the ketoacyl-synthase (KS) domain and away from other catalytic centers, such as acetyl-transferase (AT) and ER domains by steric blockage of the KS active site followed by addition of substrates. These studies strongly suggest that acylation of phosphopantetheine arm of ACP may be an integral part of the substrate shuttling mechanism in type I fungal FAS.
Organizational Affiliation:
Department of Medical Biophysics, University of Toronto, Princess Margaret Cancer Research Institute, Toronto, Ontario, Canada.