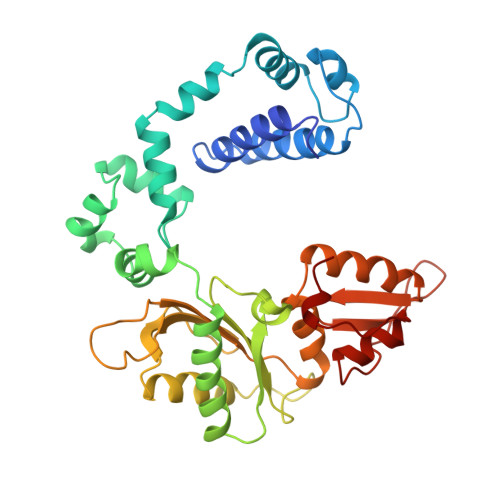
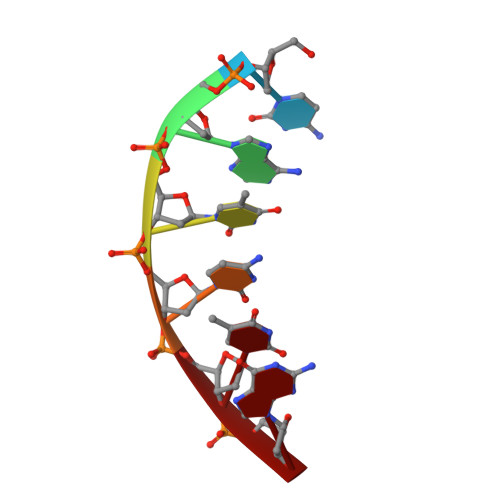
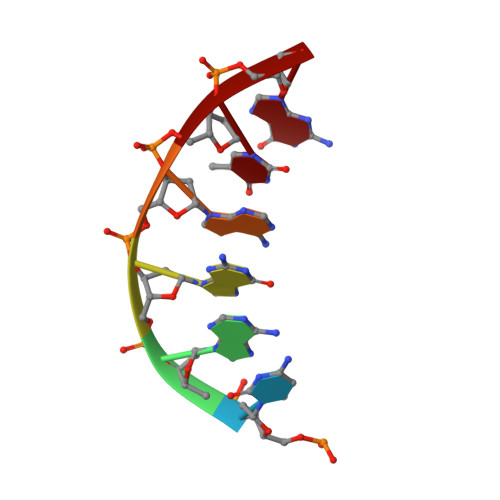
A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta
Pelletier, H., Sawaya, M.R., Wolfle, W., Wilson, S.H., Kraut, J.(1996) Biochemistry 35: 12762-12777
- PubMed: 8841119
- DOI: https://doi.org/10.1021/bi9529566
- Primary Citation of Related Structures:
1ZQT, 7ICE, 7ICF, 7ICG, 7ICH, 7ICI, 7ICJ, 7ICK, 7ICL, 7ICM, 7ICN, 7ICO, 7ICP, 7ICQ, 7ICR, 7ICS, 7ICT, 7ICU, 7ICV, 8ICJ, 8ICK, 8ICL, 8ICM, 8ICN, 8ICO, 8ICP, 8ICQ, 8ICR, 8ICS, 8ICT, 8ICU, 8ICV, 8ICW, 8ICX, 8ICY, 9ICF, 9ICK, 9ICN, 9ICO, 9ICP, 9ICQ, 9ICR, 9ICS, 9ICT, 9ICU, 9ICV - PubMed Abstract:
When crystals of human DNA polymerase beta (pol beta) complexed with DNA [Pelletier, H., Sawaya, M. R., Wolfle, W., Wilson, S. H., & Kraut, J. (1996) Biochemistry 35, 12742-12761] are soaked in the presence of dATP and Mn2+, X-ray structural analysis shows that nucleotidyl transfer to the primer 3'-OH takes place directly in the crystals, even though the DNA is blunt-ended at the active site. Under similar crystal-soaking conditions, there is no evidence for a reaction when Mn2+ is replaced by Mg2+, which is thought to be the divalent metal ion utilized by most polymerases in vivo. These results suggest that one way Mn2+ may manifest its mutagenic effect on polymerases is by promoting greater reactivity than Mg2+ at the catalytic site, thereby allowing the nucleotidyl transfer reaction to take place with little or no regard to instructions from a template. Non-template-directed nucleotidyl transfer is also observed when pol beta-DNA cocrystals are soaked in the presence of dATP and Zn2+, but the reaction products differ in that the sugar moiety of the incorporated nucleotide appears distorted or otherwise cleaved, in agreement with reports that Zn2+ may act as a polymerase inhibitor rather than as a mutagen [Sirover, M. A., & Loeb, L. A. (1976) Science 194, 1434-1436]. Although no reaction is observed when crystals are soaked in the presence of dATP and other metal ions such as Ca2+, Co2+, Cr3+, or Ni2+, X-ray structural analyses show that these metal ions coordinate the triphosphate moiety of the nucleotide in a manner that differs from that observed with Mg2+. In addition, all metal ions tested, with the exception of Mg2+, promote a change in the side-chain position of aspartic acid 192, which is one of three highly conserved active-site carboxylate residues. Soaking experiments with nucleotides other than dATP (namely, dCTP, dGTP, dTTP, ATP, ddATP, ddCTP, AZT-TP, and dATP alpha S) reveal a non-base-specific binding site on pol beta for the triphosphate and sugar moieties of a nucleotide, suggesting a possible mechanism for nucleotide selectivity whereby triphosphate-sugar binding precedes a check for correct base pairing with the template.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla 92093-0506, USA.