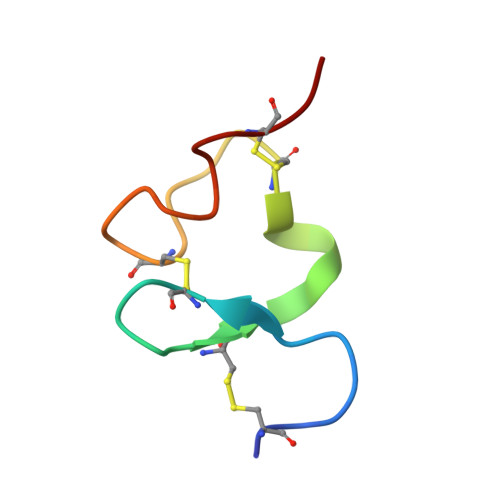
Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor.
Basore, K., Ma, H., Kafai, N.M., Mackin, S., Kim, A.S., Nelson, C.A., Diamond, M.S., Fremont, D.H.(2021) Nature 598: 672-676
- PubMed: 34646020
- DOI: https://doi.org/10.1038/s41586-021-03963-9
- Primary Citation of Related Structures:
7N1H, 7N1I - PubMed Abstract:
LDLRAD3 is a recently defined attachment and entry receptor for Venezuelan equine encephalitis virus (VEEV) 1 , a New World alphavirus that causes severe neurological disease in humans. Here we present near-atomic-resolution cryo-electron microscopy reconstructions of VEEV virus-like particles alone and in a complex with the ectodomains of LDLRAD3. Domain 1 of LDLRAD3 is a low-density lipoprotein receptor type-A module that binds to VEEV by wedging into a cleft created by two adjacent E2-E1 heterodimers in one trimeric spike, and engages domains A and B of E2 and the fusion loop in E1. Atomic modelling of this interface is supported by mutagenesis and anti-VEEV antibody binding competition assays. Notably, VEEV engages LDLRAD3 in a manner that is similar to the way that arthritogenic alphaviruses bind to the structurally unrelated MXRA8 receptor, but with a much smaller interface. These studies further elucidate the structural basis of alphavirus-receptor interactions, which could inform the development of therapies to mitigate infection and disease against multiple members of this family.
- Department of Pathology & Immunology, Washington University School of Medicine, St Louis, MO, USA.
Organizational Affiliation: