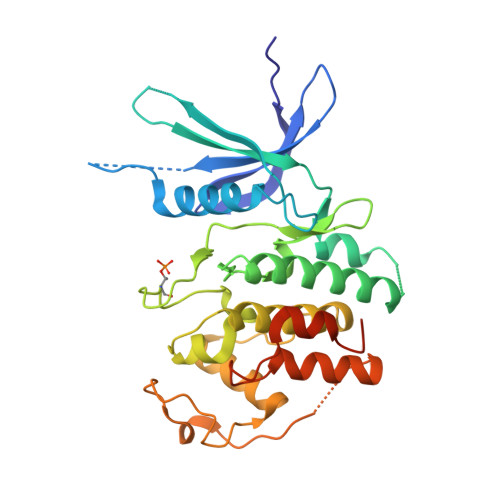
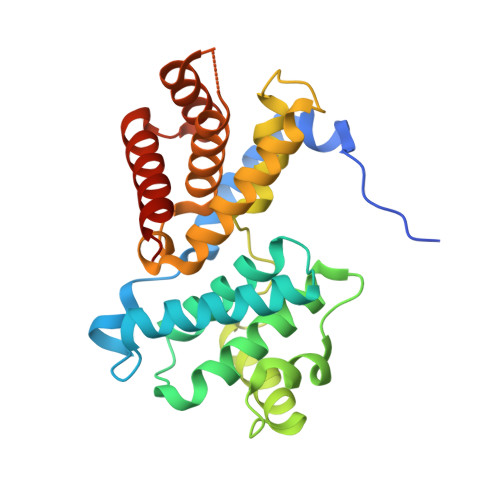
Crystal structure of active CDK4-cyclin D and mechanistic basis for abemaciclib efficacy.
Gharbi, S.I., Pelletier, L.A., Espada, A., Gutierrez, J., Sanfeliciano, S.M.G., Rauch, C.T., Ganado, M.P., Baquero, C., Zapatero, E., Zhang, A., Benach, J., Russell, A.M., Cano, L., Gomez, S., Broughton, H., Pulliam, N., Perez, C.M., Torres, R., Debets, M.F., de Dios, A., Puig, O., Hilgers, M.T., Lallena, M.J.(2022) NPJ Breast Cancer 8: 126-126
- PubMed: 36446794
- DOI: https://doi.org/10.1038/s41523-022-00494-y
- Primary Citation of Related Structures:
7SJ3 - PubMed Abstract:
Despite the biological and therapeutic relevance of CDK4/6 for the treatment of HR+, HER2- advanced breast cancer, the detailed mode of action of CDK4/6 inhibitors is not completely understood. Of particular interest, phosphorylation of CDK4 at T172 (pT172) is critical for generating the active conformation, yet no such crystal structure has been reported to date. We describe here the x-ray structure of active CDK4-cyclin D3 bound to the CDK4/6 inhibitor abemaciclib and discuss the key aspects of the catalytically-competent complex. Furthermore, the effect of CDK4/6 inhibitors on CDK4 T172 phosphorylation has not been explored, despite its role as a potential biomarker of CDK4/6 inhibitor response. We show mechanistically that CDK4/6i stabilize primed (pT172) CDK4-cyclin D complex and selectively displace p21 in responsive tumor cells. Stabilization of active CDK4-cyclin D1 complex can lead to pathway reactivation following alternate dosing regimen. Consequently, sustained binding of abemaciclib to CDK4 leads to potent cell cycle inhibition in breast cancer cell lines and prevents rebound activation of downstream signaling. Overall, our study provides key insights demonstrating that prolonged treatment with CDK4/6 inhibitors and composition of the CDK4/6-cyclin D complex are both critical determinants of abemaciclib efficacy, with implications for this class of anticancer therapy.
Organizational Affiliation:
Discovery Chemistry Research & Technology, Eli Lilly and Company, Madrid, Spain.