Crystal structure of pOmpT
Liu, J.H., Ran, T.T., Jiang, L.Y., Wang, W.W., Gao, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| OmpT | 298 | Escherichia coli | Mutation(s): 0 EC: 3.4.23.49 Membrane Entity: Yes | 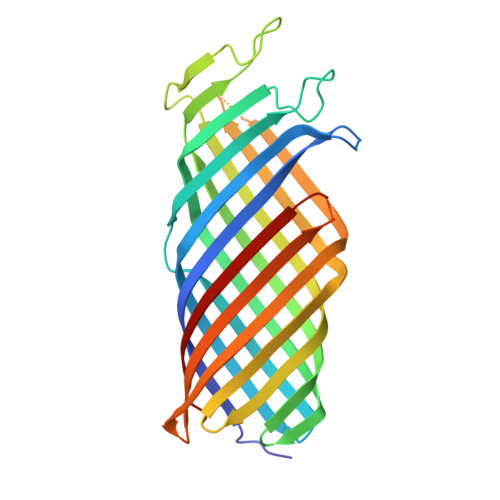 | |
UniProt | |||||
Find proteins for Q3L7I1 (Escherichia coli) Explore Q3L7I1 Go to UniProtKB: Q3L7I1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q3L7I1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 93.765 | α = 90 |
| b = 93.765 | β = 90 |
| c = 150.344 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 31972711 |
| National Natural Science Foundation of China (NSFC) | China | 31672553 |