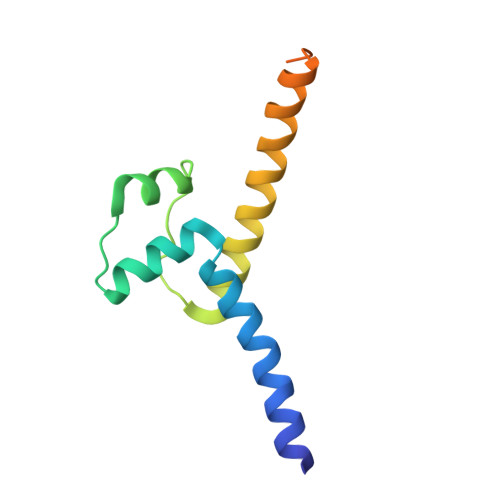
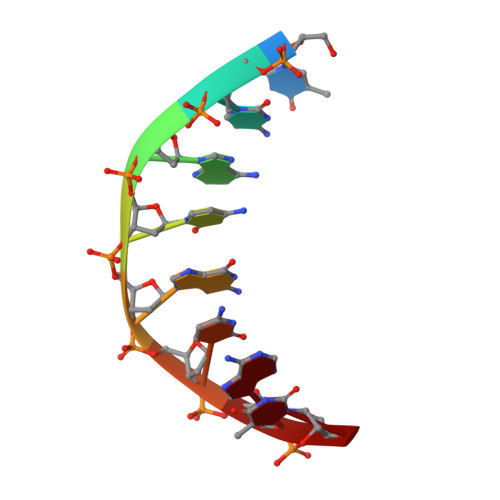
Molecular recognition of the promoter DNA signature sequence by Hms1p DBD.
Khan, M.H., Chenchen, W., Rahman, N., Lu, X., Zeng, F., Zhu, Z., Niu, L.(2025) Int J Biol Macromol : 139232-139232
- PubMed: 39756762
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.139232
- Primary Citation of Related Structures:
8HOV - PubMed Abstract:
Transcriptional regulation of sterol biosynthetic genes is mediated by conserved sterol-regulatory element binding proteins (SREBPs) in human pathogenic fungi, however, its homolog in S. cerevisiae regulate filamentous growth during stress conditions. These pseudohyphal growths might be associated with the expression of MEP2 gene in response to ammonium limitation. Hitherto, there is limited literature available for Hms1p and precisely how it establishes interaction with DNA. Though DNA and Hms1p mutual interaction was predicted computationally, however, the structural details regarding how they establish interaction still remains elusive. Here, we resolved the crystal structure of Hms1p DBD -DNA complex at a nominal resolution of 2.77 Å. The structure highlighted several residues (Hms1p His3/Asn4/Glu7/Tyr10/Arg11 ) could specifically recognize the core signature sequence in the promoter DNA fragment, which was validated by biochemical assays. Comparative analysis of Hms1p with other basic helix-loop-helix (bHLH) transcriptional regulators reflected that residues (His, Glu and Arg) are highly conserved. Despite distinct core signature sequences, these conserved residues in different bHLH proteins could specifically recognize and bind their corresponding promoter DNA fragment. Collectively, these results could pinpoint critical residues (Hms1p His3/Asn4/Glu7/Tyr10/Arg11 ) for the binding interface with the signature sequence of MEP2 promoter DNA fragment.
- MOE Key Laboratory for Membraneless Organelles and Cellular Dynamics, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China; Department of Systems Biology, School of Life Sciences, Southern University of Science and Technology, Shenzhen, China.
Organizational Affiliation: