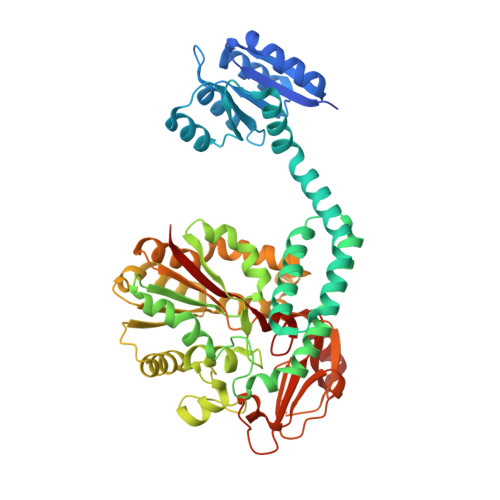
Structure-function analysis of PorX Fj , the PorX homolog from Flavobacterium johnsioniae, suggests a role of the CheY-like domain in type IX secretion motor activity.
Zammit, M., Bartoli, J., Kellenberger, C., Melani, P., Roussel, A., Cascales, E., Leone, P.(2024) Sci Rep 14: 6577-6577
- PubMed: 38503809
- DOI: https://doi.org/10.1038/s41598-024-57089-9
- Primary Citation of Related Structures:
8P6F - PubMed Abstract:
The type IX secretion system (T9SS) is a large multi-protein transenvelope complex distributed into the Bacteroidetes phylum and responsible for the secretion of proteins involved in pathogenesis, carbohydrate utilization or gliding motility. In Porphyromonas gingivalis, the two-component system PorY sensor and response regulator PorX participate to T9SS gene regulation. Here, we present the crystal structure of PorX Fj , the Flavobacterium johnsoniae PorX homolog. As for PorX, the PorX Fj structure is comprised of a CheY-like N-terminal domain and an alkaline phosphatase-like C-terminal domain separated by a three-helix bundle central domain. While not activated and monomeric in solution, PorX Fj crystallized as a dimer identical to active PorX. The CheY-like domain of PorX Fj is in an active-like conformation, and PorX Fj possesses phosphodiesterase activity, in agreement with the observation that the active site of its phosphatase-like domain is highly conserved with PorX.
- Laboratoire d'Ingénierie des Systèmes Macromoléculaires (LISM, UMR7255), Institut de Microbiologie de la Méditerranée, Aix Marseille Univ, Centre National de la Recherche Scientifique, Marseille, France.
Organizational Affiliation: