Structural insights into context-dependent inhibitory mechanisms of chloramphenicol in cells.
Xue, L., Spahn, C.M.T., Schacherl, M., Mahamid, J.(2025) Nat Struct Mol Biol 32: 257-267
- PubMed: 39668257
- DOI: https://doi.org/10.1038/s41594-024-01441-0
- Primary Citation of Related Structures:
8P6P, 8P7X, 8P7Y, 8P8B, 8P8V, 8P8W - PubMed Abstract:
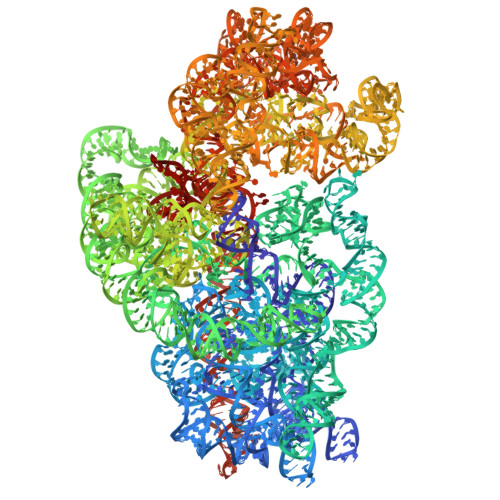
Ribosome-targeting antibiotics represent an important class of antimicrobial drugs. Chloramphenicol (Cm) is a well-studied ribosomal peptidyl transferase center (PTC) binder and growing evidence suggests that its inhibitory action depends on the sequence of the nascent peptide. How such selective inhibition on the molecular scale manifests on the cellular level remains unclear. Here, we use cryo-electron tomography to analyze the impact of Cm inside the bacterium Mycoplasma pneumoniae. By resolving the Cm-bound ribosomes to 3.0 Å, we elucidate Cm's coordination with natural nascent peptides and transfer RNAs in the PTC. We find that Cm leads to the accumulation of a number of translation elongation states, indicating ongoing futile accommodation cycles, and to extensive ribosome collisions. We, thus, suggest that, beyond its direct inhibition of protein synthesis, the action of Cm may involve the activation of cellular stress responses. This work exemplifies how in-cell structural biology can expand the understanding of mechanisms of action for extensively studied antibiotics.
- Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany. liangxue@ibp.ac.cn.
Organizational Affiliation: