Biochemical and Structural Characterisation of a Bacterial Lactoperoxidase.
Pecanac, O., Martin, C., Savino, S., Rozeboom, H.J., Fraaije, M.W., Loncar, N.(2025) Chembiochem 26: e202400713-e202400713
- PubMed: 39570012
- DOI: https://doi.org/10.1002/cbic.202400713
- Primary Citation of Related Structures:
8S6C - PubMed Abstract:
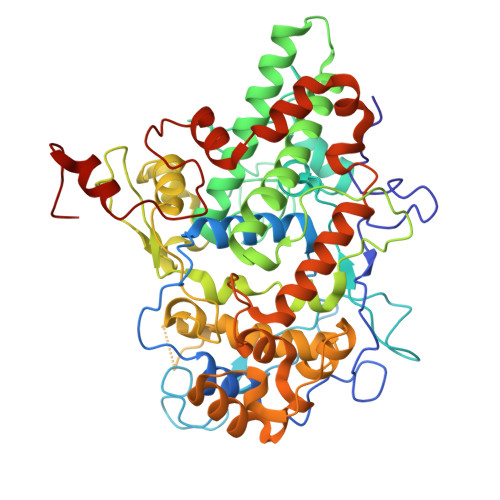
Peroxidases belong to a group of enzymes that are widely found in animals, plants and microorganisms. These enzymes are effective biocatalysts for a wide range of oxidations on various substrates. This work presents a biochemical and structural characterization of a novel heme-containing peroxidase from Cyanobacterium sp. TDX16, CyanoPOX. This cyanobacterial enzyme was successfully overexpressed in Escherichia coli as a soluble, heme-containing monomeric enzyme. Although CyanoPOX shares relatively low sequence identity (37 %) with bovine lactoperoxidase, it displays comparable biochemical properties. CyanoPOX is most stable and active in slightly acidic conditions (pH 6-6.5) and moderately thermostable (melting temperature around 48 °C). Several compounds that are typical substrates for mammalian lactoperoxidases were tested to establish the catalytic potential of CyanoPOX. Potassium iodide showed the highest catalytic efficiency (126 mM -1 s -1 ), while various aromatic compounds were also readily converted. Structural elucidation of CyanoPOX confirmed the presence of a non-covalently bound b-type heme cofactor that is situated in the central core of the protein. Except for a highly similar overall structure, CyanoPOX also has a conserved active site pocket when compared with mammalian lactoperoxidases. Due to its catalytic properties and high expression in a bacterial host, this newly discovered peroxidase shows promise for applications.
- GECCO Biotech, Zernikepark 6, 9747 AN, Groningen, The Netherlands.
Organizational Affiliation: