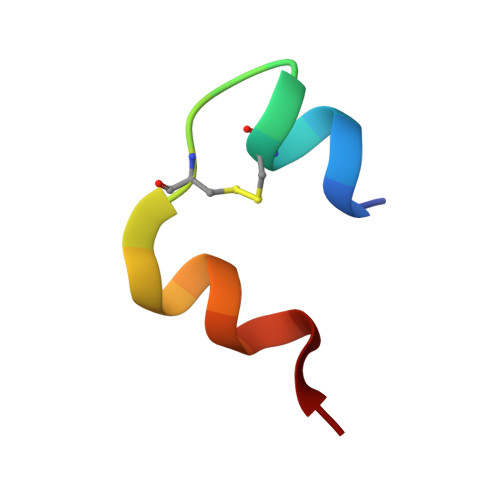
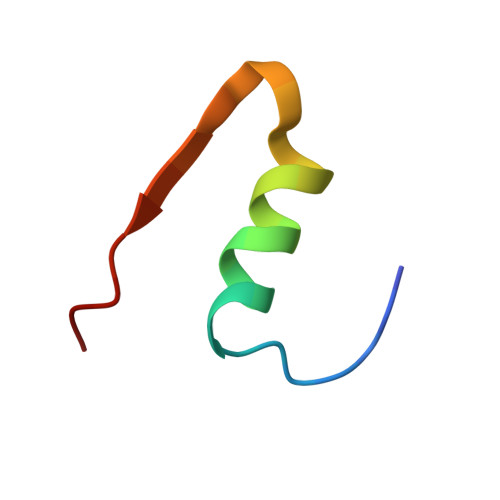
X-ray crystallographic and hydrogen deuterium exchange studies confirm alternate kinetic models for homolog insulin monomers.
Ayan, E., Turk, M., Tatli, O., Bostan, S., Telek, E., Dingiloglu, B., Dogan, B.Z., Alp, M.I., Kati, A., Dinler-Doganay, G., Demirci, H.(2025) PLoS One 20: e0319282-e0319282
- PubMed: 40257998
- DOI: https://doi.org/10.1371/journal.pone.0319282
- Primary Citation of Related Structures:
8Z4B - PubMed Abstract:
Despite the crucial role of various insulin analogs in achieving satisfactory glycemic control, a comprehensive understanding of their in-solution dynamic mechanisms still holds the potential to further optimize rapid insulin analogs, thus significantly improving the well-being of individuals with Type 1 Diabetes. Here, we employed hydrogen-deuterium exchange mass spectrometry to decipher the molecular dynamics of newly modified and functional insulin analog. A comparative analysis of H/D dynamics demonstrated that the modified insulin exchanges deuterium atoms faster and more extensively than the intact insulin aspart. Additionally, we present new insights derived from our 2.5 Å resolution X-ray crystal structure of modified hexamer insulin analog at ambient temperature. Furthermore, we obtained a distinctive side-chain conformation of the Asn3 residue on the B chain (AsnB3) by operating a comparative analysis with a previously available cryogenic rapid-acting insulin structure (PDB_ID: 4GBN). The experimental conclusions have demonstrated compatibility with modified insulin's distinct cellular activity, comparably to aspart. Additionally, the hybrid structural approach combined with computational analysis employed in this study provides novel insight into the structural dynamics of newly modified and functional insulin vs insulin aspart monomeric entities. It allows further molecular understanding of intermolecular interrelations driving dissociation kinetics and, therefore, a fast action mechanism.
- Department of Molecular Biology and Genetics, Faculty of Science, Koç University, Istanbul, Türkiye.
Organizational Affiliation: