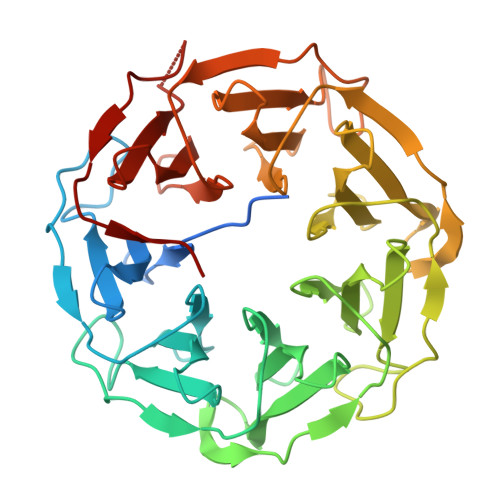
The insight into the biology of five homologous lectins produced by the entomopathogenic bacterium and nematode symbiont Photorhabdus laumondii.
Paulenova, E., Dobes, P., Melicher, F., Houser, J., Faltinek, L., Hyrsl, P., Wimmerova, M.(2025) Glycobiology
- PubMed: 40459235
- DOI: https://doi.org/10.1093/glycob/cwaf033
- Primary Citation of Related Structures:
8Q7U, 8Q80, 8Q81, 8Q82, 8Q83 - PubMed Abstract:
Photorhabdus laumondii is a well-known bacterium with a complex life cycle involving mutualism with nematodes of the genus Heterorhabditis and pathogenicity towards insect hosts. It provides an excellent model for studying the diverse roles of lectins, saccharide-binding proteins, in both symbiosis and pathogenicity. This study focuses on the seven-bladed β-propeller lectins of P. laumondii (PLLs), examining their biochemical properties (structure and saccharide specificity) and biological functions (gene expression, interactions with the nematode symbiont, and the host immune system response). Structural analyses revealed diverse oligomeric states among PLLs and a unique organisation of binding sites not described outside the PLL lectin family. Lectins exhibited high specificity for fucosylated and O-methylated saccharides with a significant avidity effect for multivalent ligands. Gene expression analysis across bacterial growth phases revealed that PLLs are predominantly expressed during the exponential phase. Interaction studies with the host immune system demonstrated that PLL5 uniquely induced melanisation in Galleria mellonella hemolymph. Furthermore, PLL2, PLL3, and PLL5 interfered with reactive oxygen species production in human blood cells, indicating their potential role in modulating host immune responses. Biofilm formation assays and binding studies with nematode life stages showed no significant involvement of PLLs in nematode colonization. Our findings highlight the primary role of PLLs in Photorhabdus pathogenicity rather than in symbiosis and offer valuable insight into the fascinating dynamics within the Photorhabdus-nematode-insect triparted system.
Organizational Affiliation:
Central European Institute of Technology (CEITEC), Masaryk University, Kamenice 5, Brno 625 00, Czech Republic.