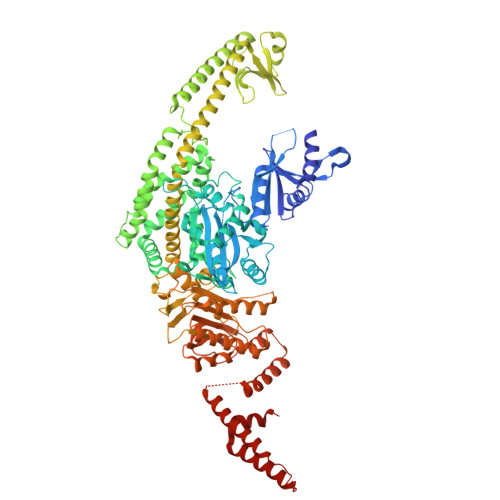
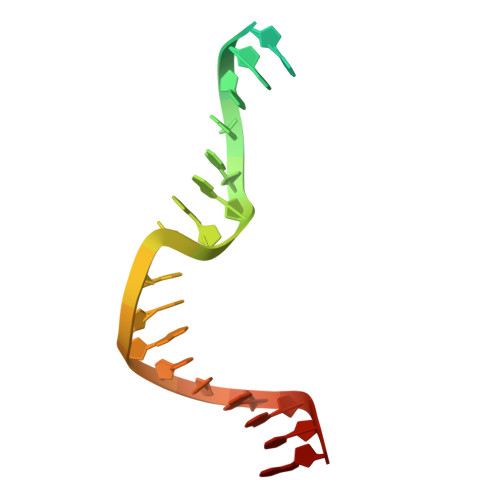
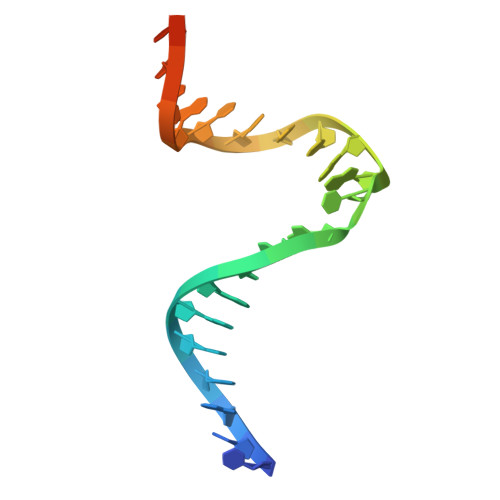
The crystal structure of DNA-bound human MutSbeta (MSH2/MSH3) in the canonical mismatch bound conformation with ADP bound in MSH2 and MSH3
Thomsen, M., Neudegger, T., Thieulin-Pardo, G., Blaesse, M., Costanzi, E., Steinbacher, S., Plotnikov, N.V., Dominguez, C., Iyer, R.R., Wilkinson, H.A., Monteagudo, E., Haque, T.S., Prasad, B.C., Finley, M., Boudet, J., Vogt, T.F., Felsenfeld, D.P.To be published.