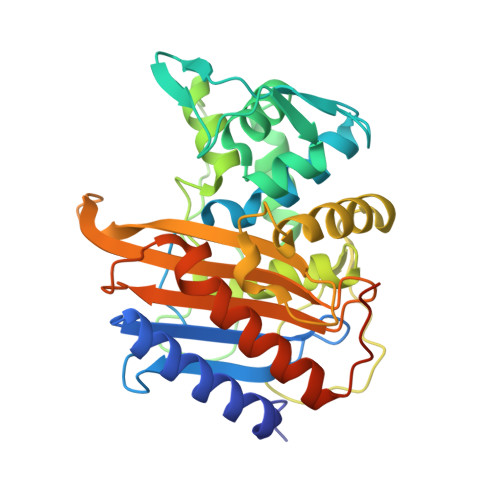
Discovery and chemical optimisation of a potent, Bi-cyclic antimicrobial inhibitor of Escherichia coli PBP3.
Rowland, C.E., Newman, H., Martin, T.T., Dods, R., Bournakas, N., Wagstaff, J.M., Lewis, N., Stanway, S.J., Balmforth, M., Kessler, C., van Rietschoten, K., Bellini, D., Roper, D.I., Lloyd, A.J., Dowson, C.G., Skynner, M.J., Beswick, P., Dawson, M.J.(2025) Commun Biol 8: 819-819
- PubMed: 40437113
- DOI: https://doi.org/10.1038/s42003-025-08246-x
- Primary Citation of Related Structures:
8RTZ - PubMed Abstract:
Penicillin binding proteins (PBPs) are well validated antimicrobial targets, but the prevalence of β-lactamase driven resistance and, more rarely, target-based mutations, necessitates new classes of PBP-targeting drugs. Here we describe the discovery and optimisation of bicyclic peptide (Bicycle ® ) inhibitors of E. coli PBP3 (EcPBP3) using a proprietary phage display platform, and their conjugation to linear antimicrobial peptides to confer outer membrane permeation. These molecules exhibited high-affinity binding to E. coli PBP3 and a viable spectrum of killing activity against clinically relevant species of the Enterobacterales. X-ray crystallography was used to explore the mode of binding to PBP3, enabling increased target affinity and improvement of in vitro stability. These compounds bind to the transpeptidase active site cleft of PBP3 and represent, to our knowledge, a novel non-β-lactam chemical class of high affinity, non-covalent penicillin binding protein inhibitors. This work demonstrates an approach to rapidly find binders to antimicrobial targets, combined with an entry mechanism to provide access to the Gram negative cell.
Organizational Affiliation:
Bicycle Tx Ltd, Blocks A&B, Portway Building, Granta Park, Great Abington, Cambridge, UK. catherine.rowland@bicycletx.com.