Integrated Study of Fluorescence Enhancement in the Y176H Variant of Cyanobacterial Phytochrome Cph1.
Nagano, S., Song, C., Rohr, V., Mackintosh, M.J., Hoang, O.T., Kraskov, A., Yang, Y., Hughes, J., Heyne, K., Mroginski, M.A., Schapiro, I., Hildebrandt, P.(2025) Biochemistry 64: 1348-1358
- PubMed: 40015976
- DOI: https://doi.org/10.1021/acs.biochem.4c00687
- Primary Citation of Related Structures:
8RVX - PubMed Abstract:
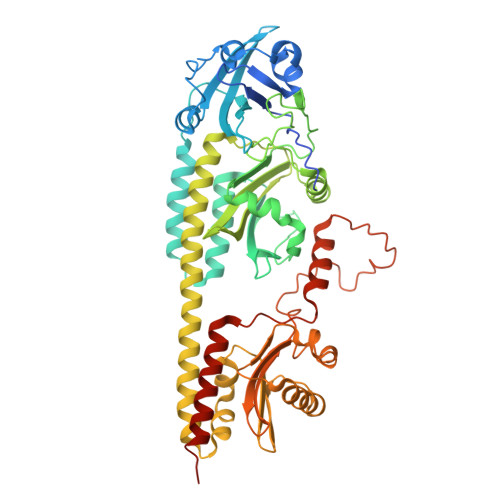
Phytochromes are red-light-sensitive biliprotein photoreceptors that control a variety of physiological processes in plants, fungi, and bacteria. Lately, greater attention has been paid to these photoreceptors due to their potential as fluorescent probes for deep-tissue microscopy. Such fluorescing phytochromes have been generated by multiple amino acid substitutions in weakly fluorescent wild-type (WT) proteins. Remarkably, the single substitution of conserved Tyr176 by His in cyanobacterial phytochrome Cph1 increases the fluorescence quantum yield from 2.4 to 14.5%. In this work, we studied this Y176H variant by crystallography, MAS NMR, resonance Raman spectroscopy, and ultrafast absorption spectroscopy complemented by theoretical methods. Two factors were identified to account for the strong fluorescence increase. First, the equilibrium between the photoactive and fluorescent substates of WT Cph1 was shown to shift entirely to the fluorescent substate in Y176H. Second, structural flexibility of the chromophore is drastically reduced and the photoisomerization barrier is raised, thereby increasing the excited-state lifetime. The most striking finding, however, is that Y176H includes the structural properties of both the dark-adapted Pr and the light-activated Pfr state. While the chromophore adopts the Pr-typical ZZZssa configuration, the tongue segment of the protein adopts a Pfr-typical α-helical structure. This implies that Tyr176 plays a key role in coupling chromophore photoisomerization to the sheet-to-helix transition of the tongue and the final Pfr structure. This conclusion extends to plant phytochromes, where the homologous substitution causes light-independent signaling activity akin to that of Pfr.
- Institute for Plant Physiology, Justus Liebig University, Senckenbergstr. 3, Giessen D-35390, Germany.
Organizational Affiliation: