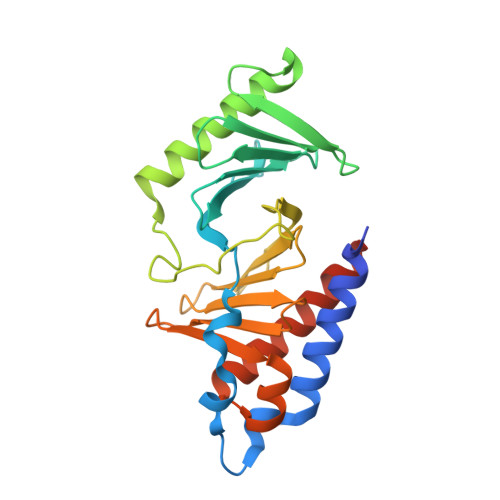
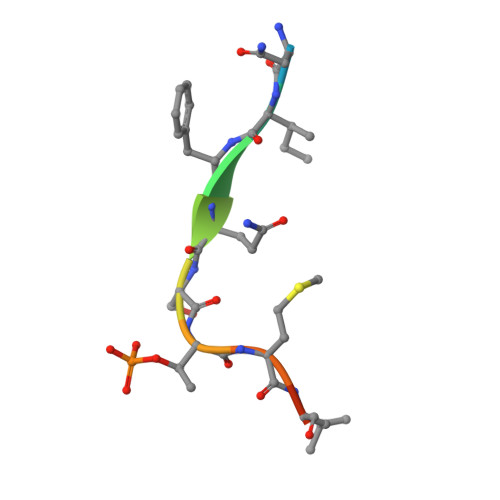
PLK1-mediated phosphorylation cascade activates Mis18 complex to ensure centromere inheritance.
Parashara, P., Medina-Pritchard, B., Abad, M.A., Sotelo-Parrilla, P., Thamkachy, R., Grundei, D., Zou, J., Spanos, C., Kumar, C.N., Basquin, C., Das, V., Yan, Z., Al-Murtadha, A.A., Kelly, D.A., McHugh, T., Imhof, A., Rappsilber, J., Jeyaprakash, A.A.(2024) Science 385: 1098-1104
- PubMed: 39236175
- DOI: https://doi.org/10.1126/science.ado8270
- Primary Citation of Related Structures:
8S30, 8S31 - PubMed Abstract:
Accurate chromosome segregation requires the attachment of microtubules to centromeres, epigenetically defined by the enrichment of CENP-A nucleosomes. During DNA replication, CENP-A nucleosomes undergo dilution. To preserve centromere identity, correct amounts of CENP-A must be restored in a cell cycle-controlled manner orchestrated by the Mis18 complex (Mis18α-Mis18β-Mis18BP1). We demonstrate here that PLK1 interacts with the Mis18 complex by recognizing self-primed phosphorylations of Mis18α (Ser 54 ) and Mis18BP1 (Thr 78 and Ser 93 ) through its Polo-box domain. Disrupting these phosphorylations perturbed both centromere recruitment of the CENP-A chaperone HJURP and new CENP-A loading. Biochemical and functional analyses showed that phosphorylation of Mis18α and PLK1 binding were required to activate Mis18α-Mis18β and promote Mis18 complex-HJURP interaction. Thus, our study reveals key molecular events underpinning the licensing role of PLK1 in ensuring accurate centromere inheritance.
- Wellcome Centre for Cell Biology, University of Edinburgh, Edinburgh EH9 3BF, UK.
Organizational Affiliation: