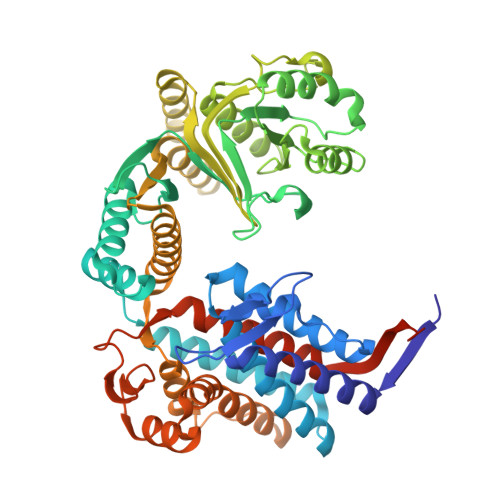
Targeted protein degradation in Escherichia coli using CLIPPERs.
Izert-Nowakowska, M.A., Klimecka, M.M., Antosiewicz, A., Wroblewski, K., Kowalski, J.J., Bandyra, K.J., Goral, T., Kmiecik, S., Serwa, R.A., Gorna, M.W.(2025) EMBO Rep 26: 3994-4016
- PubMed: 40562793
- DOI: https://doi.org/10.1038/s44319-025-00510-9
- Primary Citation of Related Structures:
8S32 - PubMed Abstract:
New, universal tools for targeted protein degradation in bacteria can help to accelerate protein function studies and antimicrobial research. We describe a new method for degrading bacterial proteins using plasmid-encoded degrader peptides which deliver target proteins for degradation by a highly conserved ClpXP protease. We demonstrate the mode of action of the degraders on a challenging essential target, GroEL. The studies in bacteria are complemented by in vitro binding and structural studies. Expression of degrader peptides results in a temperature-dependent growth inhibition and depletion of GroEL levels over time. The reduction of GroEL levels is accompanied by dramatic proteome alterations. The presented method offers a new alternative approach for regulating protein levels in bacteria without genomic modifications or tag fusions. Our studies demonstrate that ClpXP is an attractive protease for the future use in bacterial-targeted protein degradation.
- Structural Biology Group, Biological and Chemical Research Centre, Faculty of Chemistry, University of Warsaw, Warsaw, Poland.
Organizational Affiliation: