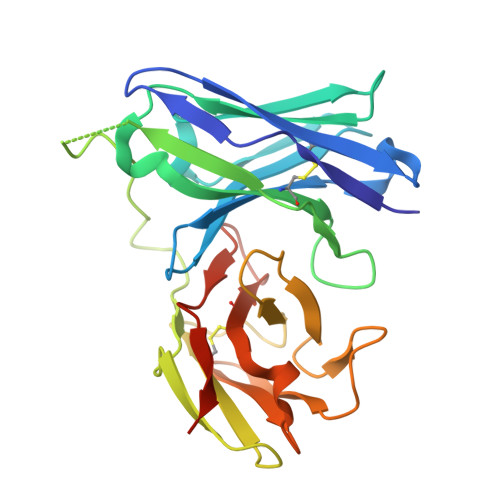
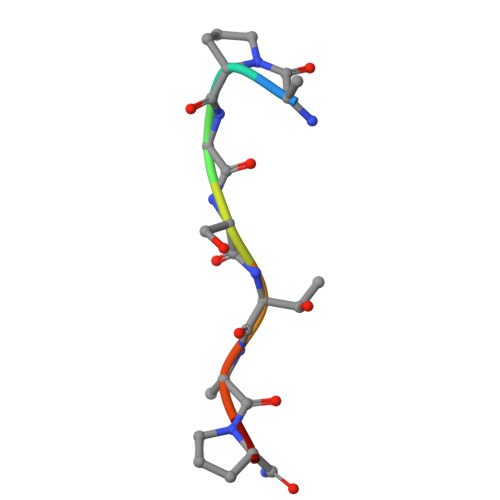
Recognizing Tn and STn Epitopes in Protein Context: Structural Advances in Phage Display Libraries for Tumor-Specific Antibody Discovery
Hurtado-Guerrero, R., Gatos, S., Gines-Alcober, I., Macias-Leon, J., Manuel Gonzalez-Ramirez, A., Kasapoglu, I., Veloz, B., Companon, I., Ghirardello, M., Merino, P., Corzana, F., Blixt, O.(2025) Nat Chem Biol