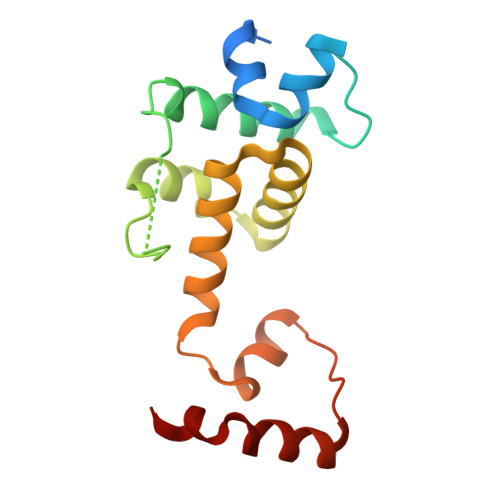
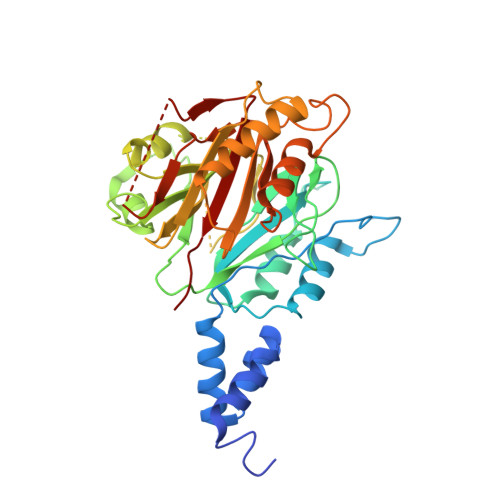
Dual BACH1 regulation by complementary SCF-type E3 ligases.
Goretzki, B., Khoshouei, M., Schroder, M., Penner, P., Egger, L., Stephan, C., Argoti, D., Dierlamm, N., Rada, J.M., Kapps, S., Muller, C.S., Thiel, Z., Mutlu, M., Tschopp, C., Furkert, D., Freuler, F., Haenni, S., Tenaillon, L., Knapp, B., Hinniger, A., Hoppe, P., Schmidt, E., Gutmann, S., Iurlaro, M., Ryzhakov, G., Fernandez, C.(2024) Cell 187: 7585-7602.e25
- PubMed: 39657677
- DOI: https://doi.org/10.1016/j.cell.2024.11.006
- Primary Citation of Related Structures:
8S7D, 8S7E, 9GP5, 9GR9, 9GRA - PubMed Abstract:
Broad-complex, tramtrack, and bric-à-brac domain (BTB) and CNC homolog 1 (BACH1) is a key regulator of the cellular oxidative stress response and an oncogene that undergoes tight post-translational control by two distinct F-box ubiquitin ligases, SCF FBXO22 and SCF FBXL17 . However, how both ligases recognize BACH1 under oxidative stress is unclear. In our study, we elucidate the mechanism by which FBXO22 recognizes a quaternary degron in a domain-swapped β-sheet of the BACH1 BTB dimer. Cancer-associated mutations and cysteine modifications destabilize the degron and impair FBXO22 binding but simultaneously expose an otherwise shielded degron in the dimer interface, allowing FBXL17 to recognize BACH1 as a monomer. These findings shed light on a ligase switch mechanism that enables post-translational regulation of BACH1 by complementary ligases depending on the stability of its BTB domain. Our results provide mechanistic insights into the oxidative stress response and may spur therapeutic approaches for targeting oxidative stress-related disorders and cancer.
- Discovery Sciences, Novartis Biomedical Research, Basel, Switzerland. Electronic address: bengoretzki@isomorphiclabs.com.
Organizational Affiliation: