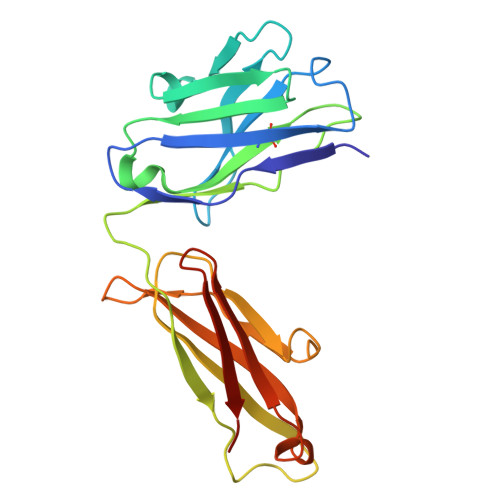
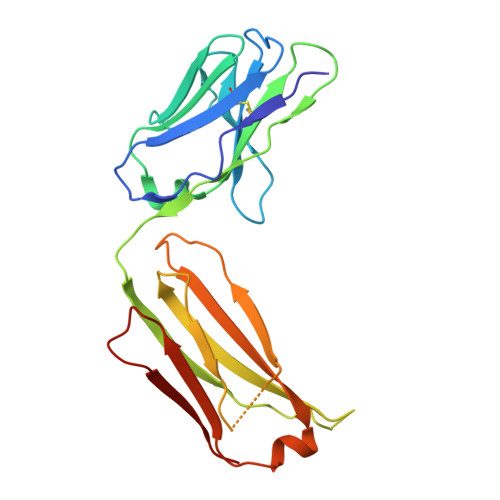
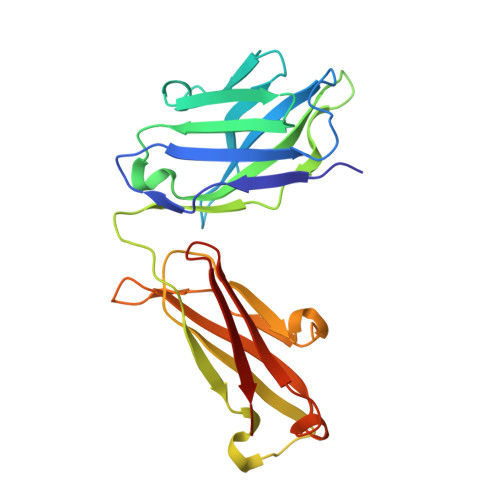
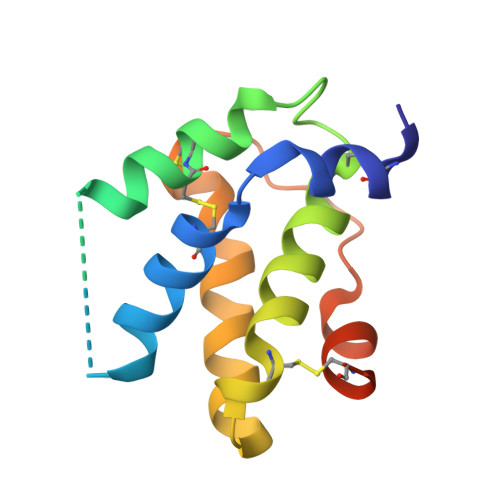
Structural determinants of peanut induced anaphylaxis.
Smith, S.A., Shrem, R.A., Lanca, B.B.C., Zhang, J., Wong, J.J.W., Croote, D., Peebles Jr., R.S., Spiller, B.W.(2025) J Allergy Clin Immunol
- PubMed: 39805366
- DOI: https://doi.org/10.1016/j.jaci.2024.12.1095
- Primary Citation of Related Structures:
8SHV, 8SHW, 8SI1, 8SJ4, 8SJ6, 8SJA - PubMed Abstract:
Human monoclonal IgE antibodies recognizing peanut allergens have recently become available, but we lack a detailed understanding of how these IgEs target allergens. To determine the molecular details of the antibody-allergen interaction for a panel of clinically important human IgE monoclonal antibodies and to develop strategies to disrupt disease causing antibody-allergen interactions. We identified candidates from a panel of epitope binned human IgE monoclonals that recognize two important and homologous peanut allergens, Ara h 2 and Ara h 6. Crystal structures were determined revealing the interfaces (antigenic sites) of exemplars of five common IgE bins. Among the common antigenic sites on Ara h 2 and Ara h 6, two sites (A and B) are highly conserved between the allergens, explaining the cross reactivity of antibodies that recognize these sites. Three sites (C, D, and F) involve residues that are not conserved between the allergens. Of the five common sites, three Sites (B, C, and D) involve residues that are only near each other when the allergens are properly folded, such that these sites are conformational. Two additional sites (sites A and F) involve largely linear stretches of amino acids. Site F targeted antibody, 38B7, binds to a peptide sequence DPYSP OH S, in which hydroxylation of the last proline is critical for binding. This sequence is repeated two or three times depending on the Ara h 2 isoform, enabling 38B7 to induce anaphylaxis as a single monoclonal, without a second antibody. We have mutated key residues in each site and created a panel of hypoallergens, having reduced IgE mAb binding and lacking the ability to induce anaphylaxis in our murine model. We created a structural map of the IgE antibody response to the most important peanut allergen proteins to enable the design of new allergy immunotherapies and vaccines.
- Department of Medicine, Vanderbilt University Medical Center, Vanderbilt University, Nashville, TN; Department of Pathology, Microbiology and Immunology, Vanderbilt University Medical Center, Vanderbilt University, Nashville, TN.
Organizational Affiliation: