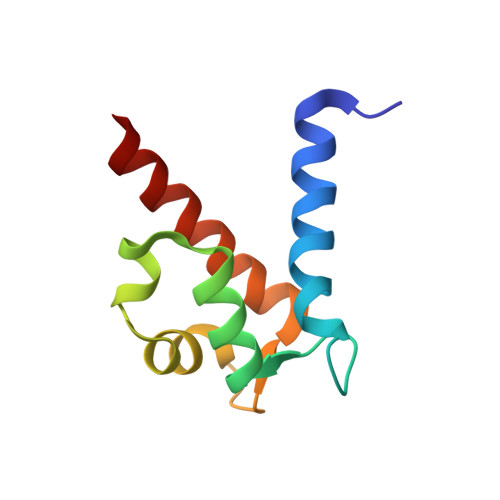
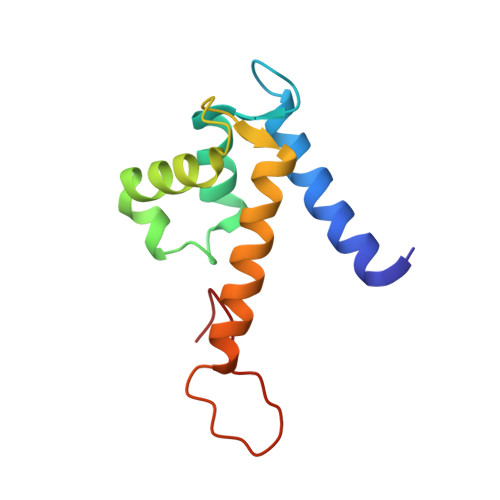
The C-terminal extension of calprotectin mediates zinc chelation and modulates Staphylococcus aureus biomass accumulation.
Perera, Y.R., Enriquez, K.T., Rodriguez, A., Garcia, V., Akizuki, T., Naretto, A., Togashi, M., Guillen, R., Skaar, E.P., Chazin, W.J.(2025) Protein Sci 34: e70294-e70294
- PubMed: 40944454
- DOI: https://doi.org/10.1002/pro.70294
- Primary Citation of Related Structures:
8SJC - PubMed Abstract:
Calprotectin (CP) is an S100A8/S100A9 heterodimer that plays an important role in nutritional immunity at the host-microbe interface. CP combats Staphylococcus aureus growth by sequestration of zinc and other trace transition metals; however, questions remain about whether CP antimicrobial activity strictly relies on metal sequestration. Moreover, the precise mechanism for how zinc binds at the two distinct transition metal binding sites of CP is not known. High-resolution X-ray crystal structures reveal tetracoordinate binding in the canonical His 3 Asp site and hexacoordinate binding in the His 6 site similar to the binding of manganese and nickel in this site. The S100A9 C-terminal extension (tail) contributes two of the His residues in the His 6 metal-binding site, but measurements of zinc affinity show there is no significant reduction upon mutation of these His residues or deletion of the entire C-terminal tail. Bacterial growth and static biofilm assays show that the His mutations affect S. aureus biomass accumulation differently than loss of the S100A9 C-terminal tail, despite resulting in the same defect in bacterial-CP binding. These results reveal that the S100A9 tail of CP has a role in preventing S. aureus biomass accumulation.
- Departments of Biochemistry and Chemistry, Vanderbilt University, Nashville, Tennessee, USA.
Organizational Affiliation: