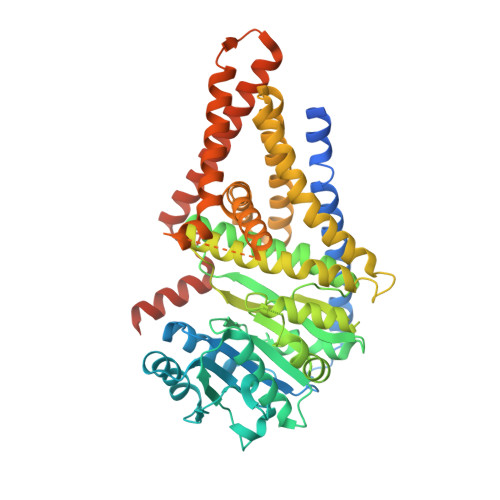
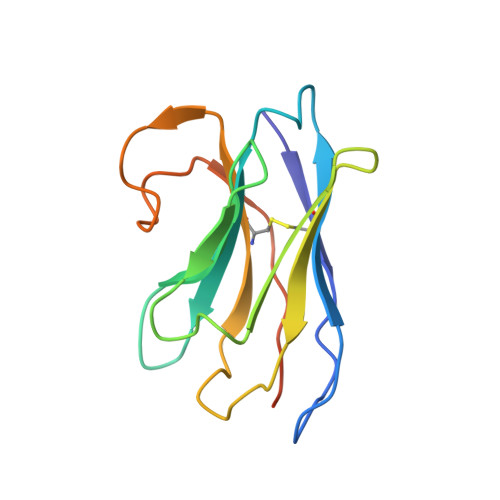
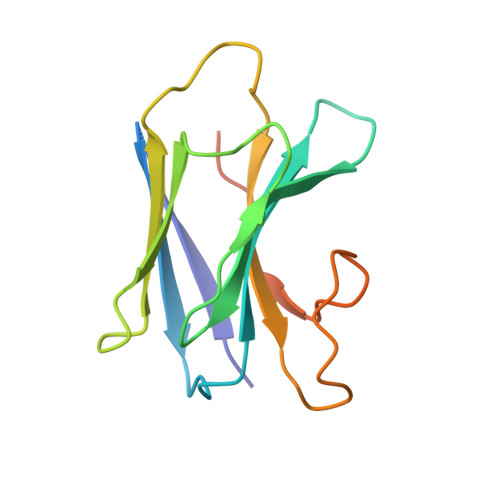
Structural insights into translocation and tailored synthesis of hyaluronan.
Gorniak, I., Stephens, Z., Erramilli, S.K., Gawda, T., Kossiakoff, A.A., Zimmer, J.(2025) Nat Struct Mol Biol 32: 161-171
- PubMed: 39322765
- DOI: https://doi.org/10.1038/s41594-024-01389-1
- Primary Citation of Related Structures:
8SMM, 8SMN, 8SMP, 8SNC, 8SND, 8SNE - PubMed Abstract:
Hyaluronan (HA) is an essential component of the vertebrate extracellular matrix. It is a heteropolysaccharide of N-acetylglucosamine (GlcNAc) and glucuronic acid (GlcA) reaching several megadaltons in healthy tissues. HA is synthesized and translocated in a coupled reaction by HA synthase (HAS). Here, structural snapshots of HAS provide insights into HA biosynthesis, from substrate recognition to HA elongation and translocation. We monitor the extension of a GlcNAc primer with GlcA, reveal the coordination of the uridine diphosphate product by a conserved gating loop and capture the opening of a translocation channel to coordinate a translocating HA polymer. Furthermore, we identify channel-lining residues that modulate HA product lengths. Integrating structural and biochemical analyses suggests an avenue for polysaccharide engineering based on finely tuned enzymatic activity and HA coordination.
- Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, VA, USA.
Organizational Affiliation: