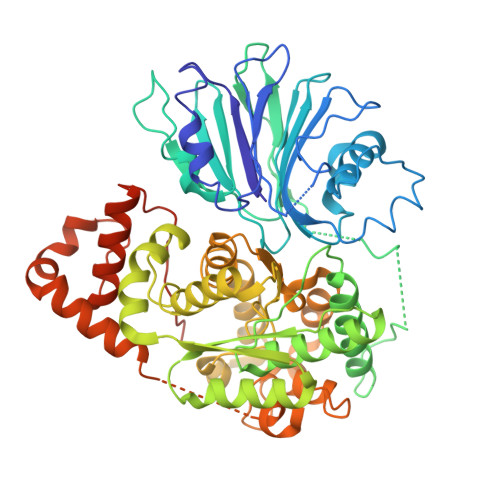
3D variability analysis reveals a hidden conformational change controlling ammonia transport in human asparagine synthetase.
Coricello, A., Nardone, A.J., Lupia, A., Gratteri, C., Vos, M., Chaptal, V., Alcaro, S., Zhu, W., Takagi, Y., Richards, N.G.J.(2024) Nat Commun 15: 10538-10538
- PubMed: 39627226
- DOI: https://doi.org/10.1038/s41467-024-54912-9
- Primary Citation of Related Structures:
8SUE, 9B6C - PubMed Abstract:
Advances in X-ray crystallography and cryogenic electron microscopy (cryo-EM) offer the promise of elucidating functionally relevant conformational changes that are not easily studied by other biophysical methods. Here we show that 3D variability analysis (3DVA) of the cryo-EM map for wild-type (WT) human asparagine synthetase (ASNS) identifies a functional role for the Arg-142 side chain and test this hypothesis experimentally by characterizing the R142I variant in which Arg-142 is replaced by isoleucine. Support for Arg-142 playing a role in the intramolecular translocation of ammonia between the active site of the enzyme is provided by the glutamine-dependent synthetase activity of the R142 variant relative to WT ASNS, and MD simulations provide a possible molecular mechanism for these findings. Combining 3DVA with MD simulations is a generally applicable approach to generate testable hypotheses of how conformational changes in buried side chains might regulate function in enzymes.
- Dipartimento di Scienze della Salute, Università "Magna Græcia" di Catanzaro, Catanzaro, Italy.
Organizational Affiliation: