The 2.3 angstrom Structure of A21, a Protein Component of the Conserved Poxvirus Entry-Fusion Complex.
Diesterbeck, U.S., Muslinkina, L.A., Gittis, A.G., Singh, K., Moss, B., Garboczi, D.N.(2025) J Mol Biology 437: 169097-169097
- PubMed: 40118206
- DOI: https://doi.org/10.1016/j.jmb.2025.169097
- Primary Citation of Related Structures:
8U0R - PubMed Abstract:
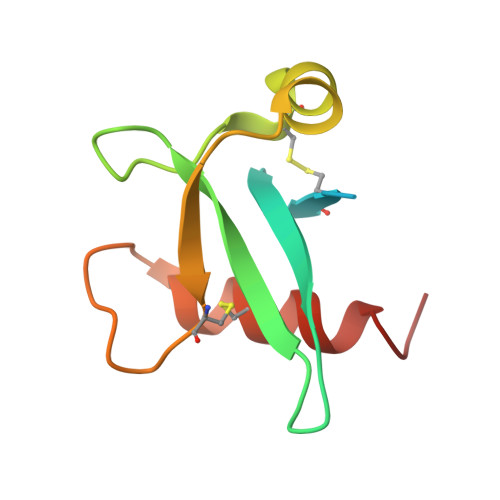
Poxviruses are exceptional in having an entry-fusion complex (EFC) consisting of eleven conserved proteins embedded in the membrane of mature virions. With the goal of understanding the function of the EFC, extensive efforts have been made to determine the structures and roles of its components, and to date, structures have been determined for nine of the eleven proteins. Here, we report the crystal structure of A21, the 10th EFC protein, comprising two α-helices clasping a twisted antiparallel β-sheet stabilized by two conserved disulfide bonds. The stability of each of the three A21 loops is provided by hydrogen bonds between main-chain atoms and several highly conserved residues, making the overall fold of A21 and its orthologs resilient to evolutionary change. Based on AlphaFold modeling and phylogenetic analysis of A21, we suggest that its highly conserved N-terminal transmembrane domain and C-terminal α-helix enable A21 integration into EFC, where it primarily interacts with the G3/L5 subcomplex and the smallest of EFC components, the O3 protein.
Organizational Affiliation:
Laboratory of Viral Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA; Ceva Innovation Center GmbH, Am Pharmapark, 06861 Dessau-Rosslau, Germany.