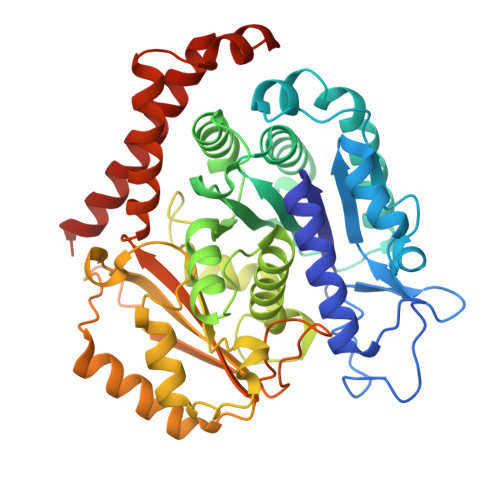
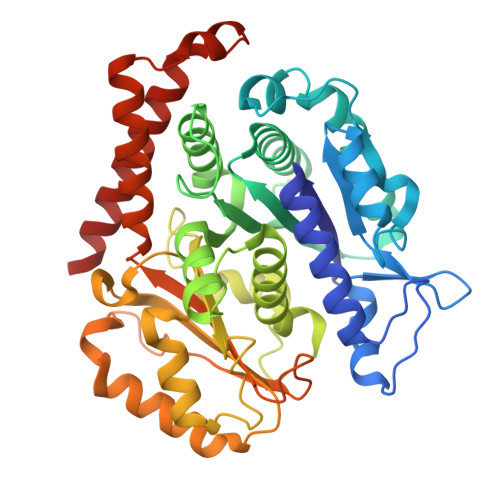
HURP binding to the vinca domain of beta-tubulin accounts for cancer drug resistance.
Saju, A., Chen, P.P., Weng, T.H., Tsai, S.Y., Tanaka, A., Tseng, Y.T., Chang, C.C., Wang, C.H., Shimamoto, Y., Hsia, K.C.(2024) Nat Commun 15: 8844-8844
- PubMed: 39397030
- DOI: https://doi.org/10.1038/s41467-024-53139-y
- Primary Citation of Related Structures:
8X9P - PubMed Abstract:
Vinca alkaloids, a class of tubulin-binding agent, are widely used in treating cancer, yet the emerging resistance compromises their efficacy. Hepatoma up-regulated protein (HURP), a microtubule-associated protein displaying heightened expression across various cancer types, reduces cancer cells' sensitivity to vinca-alkaloid drugs upon overexpression. However, the molecular basis behind this drug resistance remains unknown. Here we discover a tubulin-binding domain within HURP, and establish its role in regulating microtubule growth. Cryo-EM analysis reveals interactions between HURP's tubulin-binding domain and the vinca domain on β-tubulin -- the site targeted by vinca alkaloid drugs. Importantly, HURP competes directly with vinorelbine, a vinca alkaloid-based chemotherapeutic agent, countering microtubule growth defects caused by vinorelbine both in vitro and in vivo. Our findings elucidate a mechanism driving drug resistance in HURP-overexpressing cancer cells and emphasize HURP tubulin-binding domain's role in mitotic spindle assembly. This underscores its potential as a therapeutic target to improve cancer treatment.
- Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan.
Organizational Affiliation: